

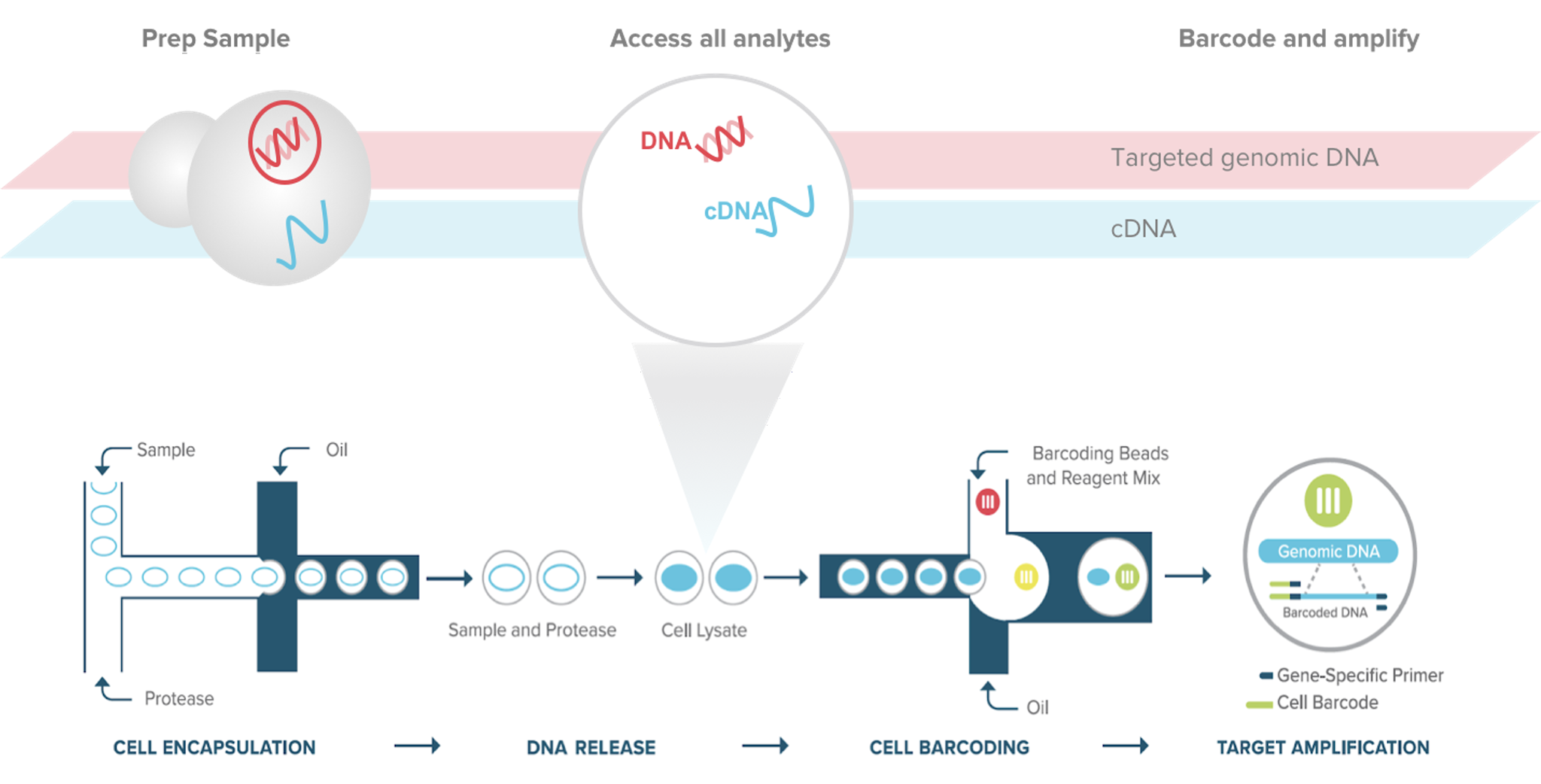
The Tapestri® Single-Cell Targeted DNA + RNA Assay delivers a multidimensional view of each cell, linking genotype to phenotype to reveal how genetic variation shapes RNA expression. From complex cancers to engineered cell therapies, Tapestri empowers researchers to uncover mechanisms of action, validate therapeutic edits, and accelerate the path to precision medicine.
By integrating targeted DNA genotyping with RNA expression profiling, you can confidently pinpoint disease-driving variants, uncover resistance mechanisms, and characterize engineered cell populations — all from the same cells.
Combine somatic mutation and expression profiles to distinguish subclones, identify driver mutations, and reveal dysregulated pathways driving disease progression.
Link genomic variants to functional RNA readouts to confirm target expression, uncover actionable biomarkers, and validate therapeutic mechanisms.
Correlate gene editing outcomes and vector copy number with transcriptional changes to verify on-target activity, assess efficacy, and ensure functional validation.
Tumor Classification & Clonal Subtyping
Identifying Driver Mutations & Pathway Dysregulation
Predicting Therapy Response
Drug Target Discovery & Biomarker Development
Rare Genetic & Inherited Disorders
Therapy Response & Mechanism of Action

This experiment highlights the power of Tapestri and its ability to distinguish cell types in cell lines and PBMCs. T47D (breast cancer) and A549 (lung cancer) were chosen to highlight the variable genotypic profiles of each disease, while PBMCs were included to show how granularly the cell types within the sample type could be distinguished and categorized.
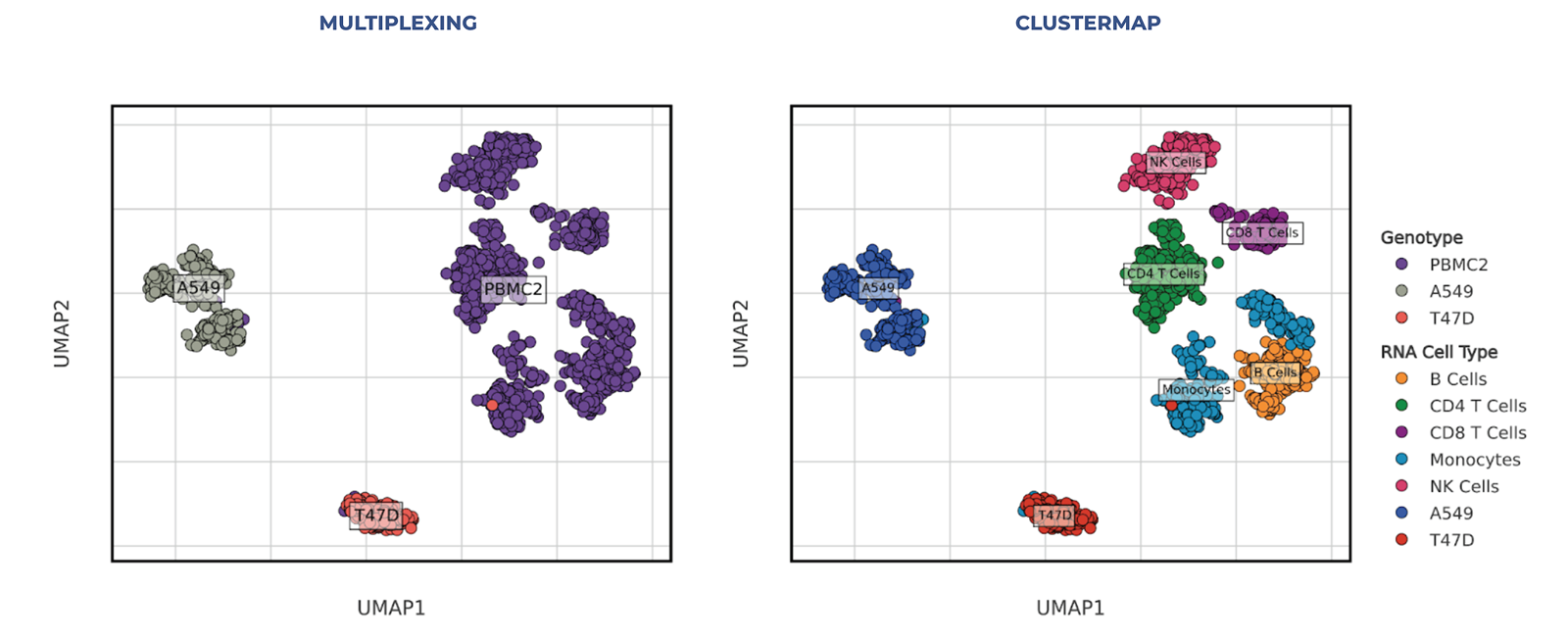
Figure 1: The ‘Genotype’ subplot displays cells from the multiplexed run, colored according to their genotype-level information derived from genotype demultiplexing, which identifies samples based on genotype variants. The ‘RNA Cell Type’ subplot shows the same cells colored by clustering results obtained from gene expression profiles of the targeted gene panel or probes.
