

Though versatile, bulk sequencing provides a blurred view of cancer’s genetic landscape, obscuring the details that could make or break research directions and treatment strategies. Single-cell multiomic analysis brings those details into sharp focus, offering a powerful tool for understanding and combating clonal heterogeneity.
Here, we discuss the limitations of traditional cancer profiling methods and explore how a multiomic approach to single-cell analysis offers a powerful alternative. By providing a comprehensive overview of clonal architecture, this high-resolution method has profound implications for diagnosis, prognosis, and treatment strategy.

As cancer progresses, mutations accumulate–leading to subpopulations, or clones, with distinct genomes. This process, known as clonal evolution, allows subpopulations with selective advantages to outcompete others.
Clonal evolution underpins clonal heterogeneity (Fig. 1). Within a single patient, cancer cell populations can exhibit significant functional and phenotypic variation. Understanding this clonal heterogeneity is crucial for prognoses and treatment strategies. However, traditional methods are complex and costly.
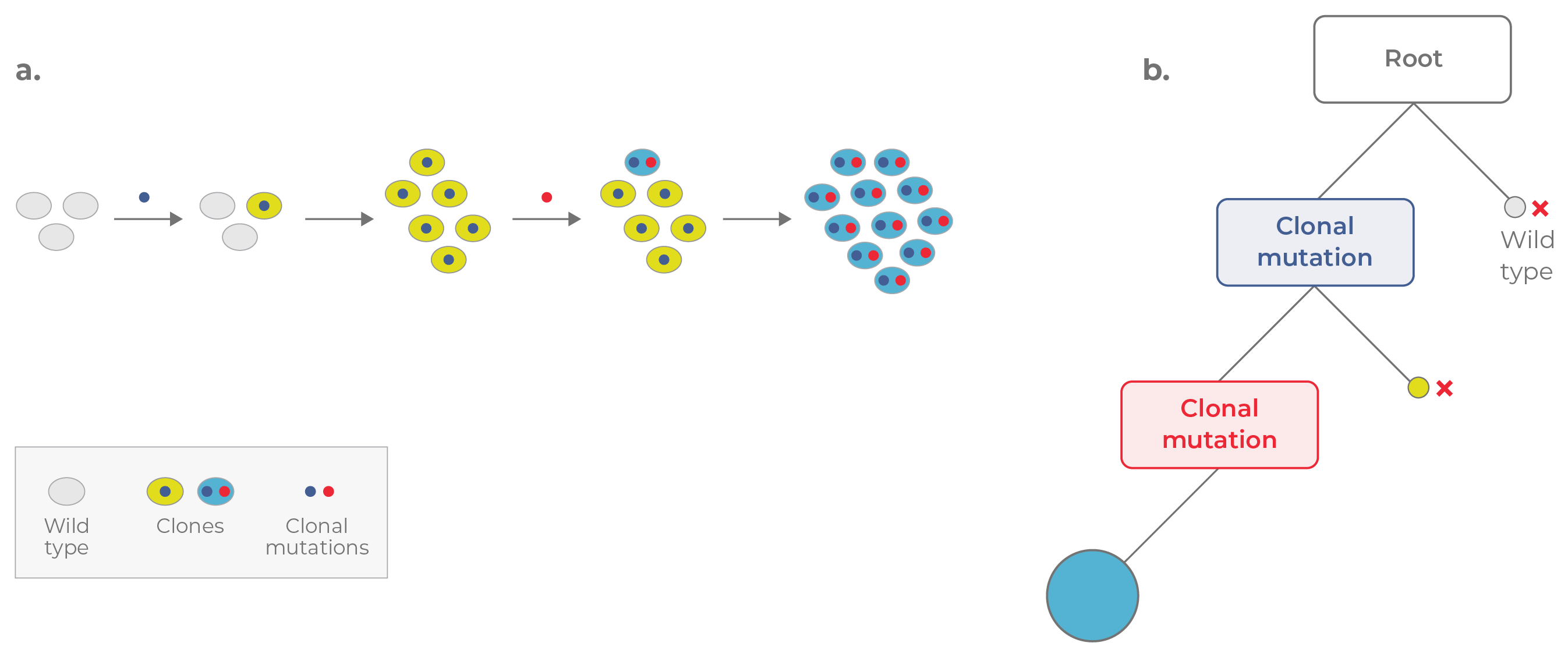
Figure 1. Linear evolution. a. In linear evolution, each mutation confers a strong selective advantage, enabling each clone to replace the clone preceding it. b. The corresponding phylogenetic tree appears linear. Mutations are acquired in a cumulative fashion, with the most recent clone (blue circle) containing all of the mutations acquired previously.
Higher clonal diversity is associated with greater disease severity, so recognizing and investigating these intrapatient differences is important for preclinical and clinical decision-making.
Genomic tumor profiling reveals clonal heterogeneity that can affect the growth and metastatic potential of tumors.1
For example, different subclones within single breast tumors may respond differently to hormone therapies, targeted treatments, or chemotherapy.2,3 Identifying these subclones will enable clinicians to prescribe combination therapies that successfully treat the entire tumor and adapt treatments as the tumor’s genetic landscape alters.
Clonal heterogeneity plays a significant role in disease progression, treatment response, and disease relapse in blood cancers.4,5
Some heme malignancies, such as multiple myeloma (MM), are especially genetically diverse, causing them to have high relapse rates.6 Understanding the complex clonal composition of a patient’s MM from diagnosis and beyond helps clinicians select the most effective treatment regimen and monitor for the emergence of resistant clones.

Figure 2. Traditional cancer profiling approaches assess a range of cellular attributes, including clonal architecture. Although bulk sequencing is versatile, it only infers clonal heterogeneity at a population level. Traditional single-cell analyses provide higher resolution but are limited to focusing on specific cellular attributes in isolation.
Standard methods for investigating clonal heterogeneity include bulk sequencing and flow cytometry (Fig. 2).7 Although these techniques have been instrumental in advancing the understanding of cancer genomics, they are often limited to focusing on isolated cellular attributes and unable to provide insight into clonal heterogeneity.
Bulk sequencing, such as next-generation sequencing (NGS), averages genetic information across all cells in a sample, preventing the identification of subclones and their specific mutations. NGS is also susceptible to inaccurate profiling when dominant clones mask less frequent but potentially crucial subclonal mutations.
These limitations result in an incomplete understanding of cancer’s genetic diversity. This is associated with an increased likelihood of selecting unsuitable therapies that don’t target the entire cancer population, inadvertently promoting the growth of resistant clones.
Many single-cell cancer profiling assays focus on specific aspects of cellular biology (e.g., immunophenotype, zygosity) and lack multiomic capabilities. Therefore, researchers must perform multiple assays. Not only is this costly, but it exhausts precious samples, leaving little material for downstream analyses.
This approach requires complex integration of data obtained from separate assays to achieve a global view of clonal heterogeneity. Harmonizing datasets from distinct assays typically requires sophisticated computational methods and bioinformatics expertise to ensure accurate alignment and reliable interpretation.
Comprehensive multiomic analysis of single cells addresses the challenges associated with traditional cancer profiling methods by offering a holistic view of clonal heterogeneity.
This approach provides a high-resolution and integrated understanding of cancer biology by simultaneously analyzing multiple molecular modalities—such as DNA, RNA, and proteins—within individual cells (Fig. 3).

Figure 3. Multiomic analysis of single cells simultaneously captures clonal heterogeneity at the genomic, transcriptomic, and proteomic levels. Bulk-sequencing is lower resolution, facilitating only population-level insights.
Given these multifaceted advantages, it is crucial to further demonstrate the clinical research utility of a multiomic approach to single-cell analysis.
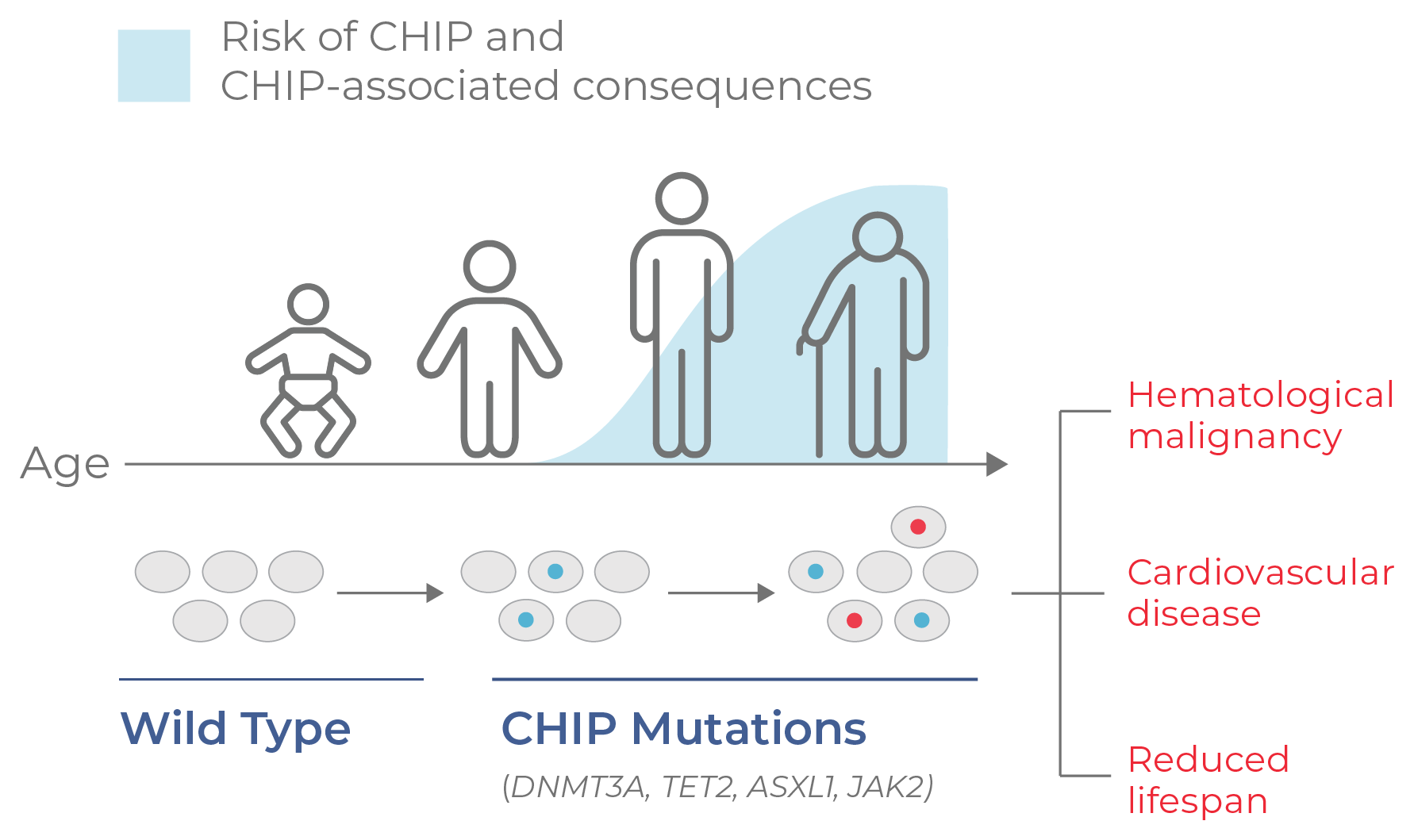
Figure 4. CHIP progresses with age. CHIP is a non-malignant condition characterized by mutation and clonal expansion of blood cells, beginning at middle age. CHIP is associated with a higher risk of developing hematological malignancies and cardiovascular disease, as well as a reduced lifespan.
Applying a multiomic approach at single-cell resolution can significantly enhance heme malignancy and genomic tumor profiling by providing detailed and integrated insights into complex cancer biology. Robust data and validation processes are essential to prove its utility and value as a clinical research tool.
Demonstrating the benefits of multiomics at the single-cell level in clinical research involves conducting retrospective and prospective studies to assess how this technology aids understanding of clonal architecture to enhance the prediction of disease progression, biomarker insights, and therapeutic target identification.
Comparative studies that showcase the increased sensitivity of multiomic analysis compared to current single-cell methods (e.g., flow cytometry for acute myeloid leukemia) will highlight the advantages of this approach in predicting disease progression, therapeutic response, and relapse.
Ultimately, continuous monitoring and documentation of the performance and outcomes of single-cell multiomic analysis in real-world translational and clinical research will further substantiate its benefits, paving the way for its widespread adoption.
For Research Use Only. Not for use in diagnostic procedures.
