

Introducing the Tapestri Single-cell Multiple Myeloma Multiomics Solution
Numerous genomic events and complex clonality in multiple myeloma (MM) render it a challenging disease with a high relapse rate despite advancements in therapies and diagnostic approaches.
Tapestri single-cell multiomics analysis transforms how you understand the complexities of multiple myeloma and its precursor stages by integrating genomic, proteomic, and clonotypic subclonal assessment with single-cell resolution, all within a single assay.
With the Tapestri Single-cell Multiple Myeloma Multiomics Solution, you gain crucial insights into the clonal and subclonal heterogeneity underlying myeloma progression, therapy response, and relapse often missed by traditional bulk methods, potentially improving the identification of novel biomarkers and the development of better therapies.
Assess clonal heterogeneity & evolution in multiple myeloma (includes SNV and focal CNA)
Discern zygosity of detected mutations impacting disease progression or drug resistance
Analyze whole genome copy number gains and losses
Identify Ig clonotype in the CDR3 region
Classify cell types and identify immunotherapy targets such as BCMA and FcRH5
Learn about our cell therapy characterization solution.

Learn how to identify smoldering multiple myeloma (SMM) clonal populations progressing to active myeloma or resistant disease with a true multiomics platform that can quantitatively correlate multiple features from SMM subclones including SNVs, genome-wide CNVs, Ig clonotype, and surface protein expression.
Featuring Adam Sciambi, PhD
CTO, Mission Bio
Tapestri automatically generates comprehensive and intuitive reports with data visualizations to help you easily correlate various clonal and subclonal attributes.
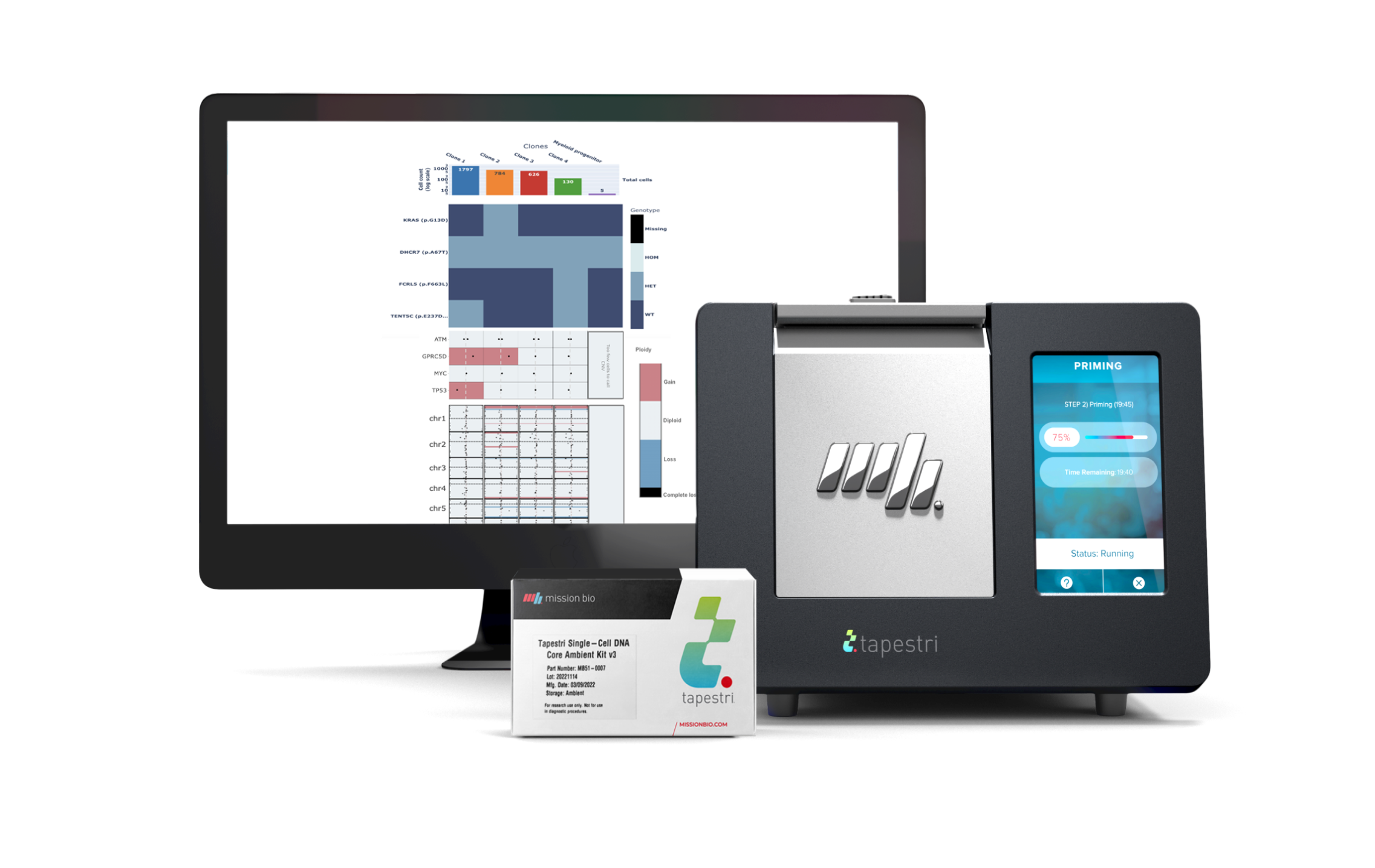
• Coverage of key driver and resistance genes, genome-wide copy number variations (CNVs), V(D)J clonotype, and cell lineage & immunotherapy markers
• Available to purchase as a complete solution or as standalone panels
500 amplicon panel uniformly covering nearly the entire genome.
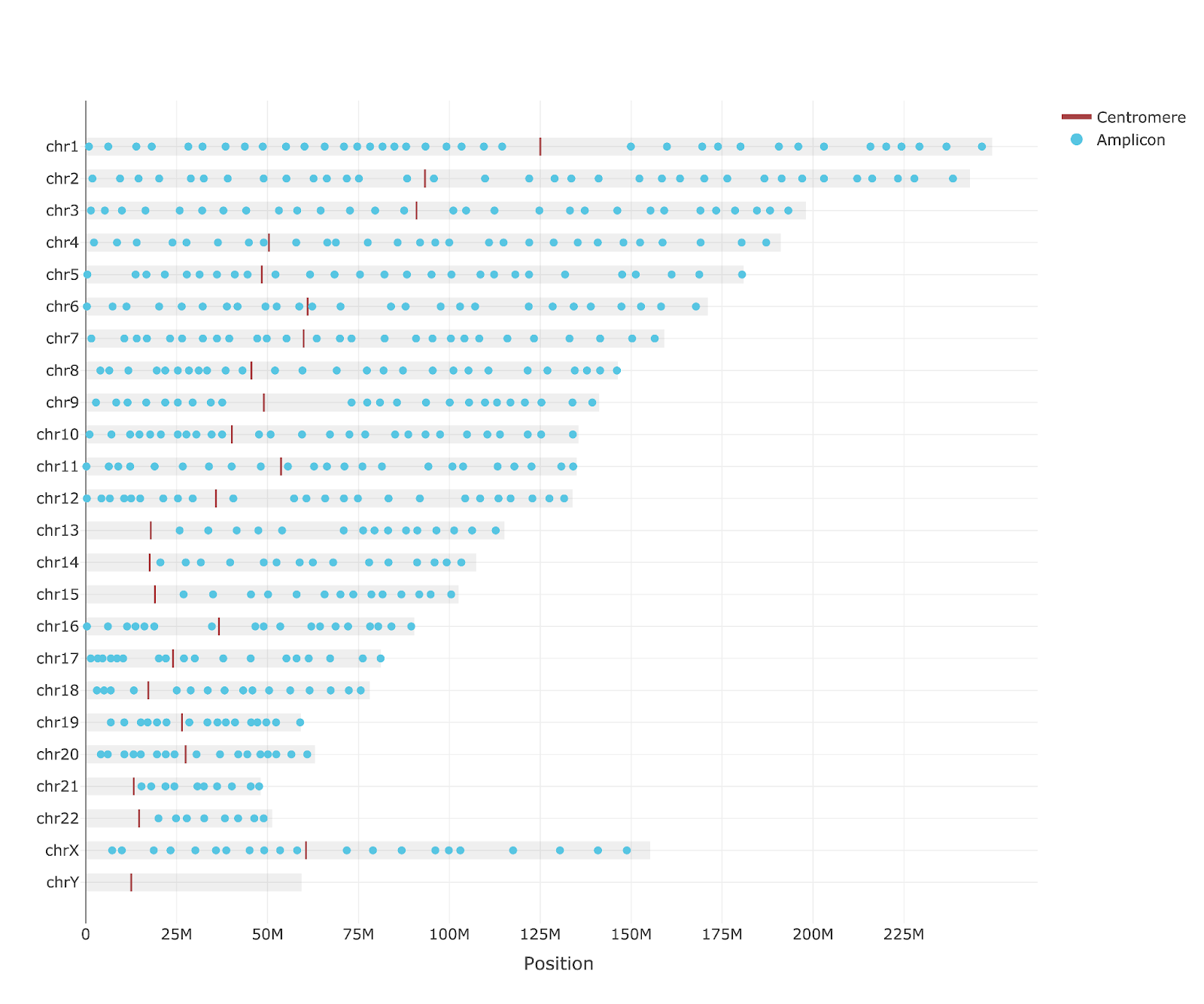
Surface antigen makers for cell type classification and therapeutic target identification.
Emerging targets for immunotherapy covered by our assay
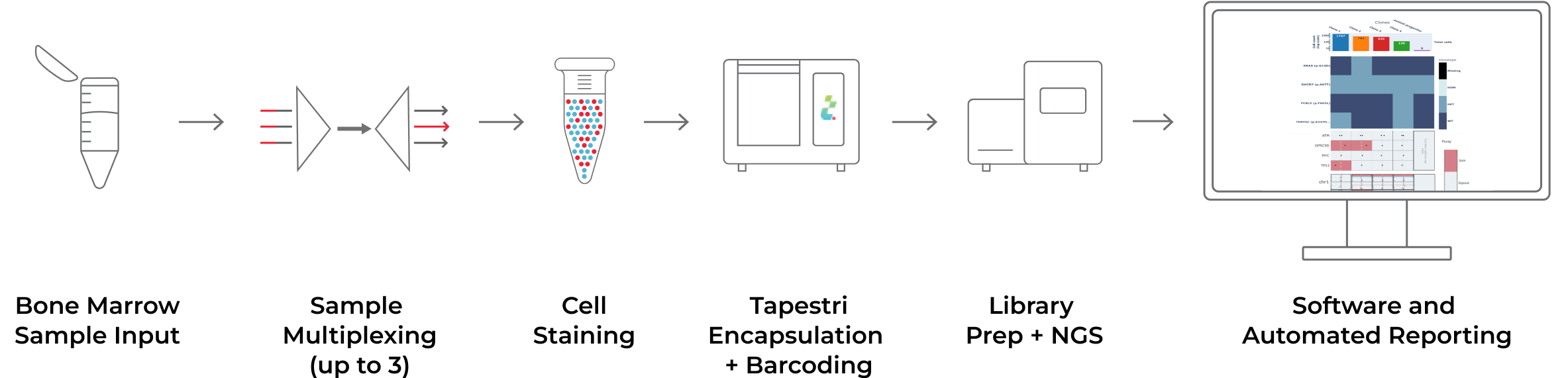
Our streamlined multiple myeloma workflow allows you to multiplex bone marrow samples on the Tapestri Platform, sequence the libraries on an NGS platform, and then easily analyze the results using the Tapestri Multiple Myeloma Software. The Solution can integrate SNVs, focal CNAs, genome-wide CNVs, V(D)J clonotype, and immunophenotype.
To support you and your drug development, we provide full service as a strategic partner for you from single-cell assay design, sample processing, through to data analysis and visualization. Whether we offer services internally or in partnership with qualified CROs, we are dedicated to the accelerated success of your drug development programs.
 LEARN MORE ABOUT PAD
LEARN MORE ABOUT PAD
Contact us to begin your evaluation
For Research Use Only. Not for use in diagnostic procedures.
