Why Single Cell?
Over the past decade, numerous advances in biotechnology have translated to improvements in the medical arena. Yet, limitations in analytical tools still remain and present significant roadblocks to the study of pathologies and the development of life-changing therapies.
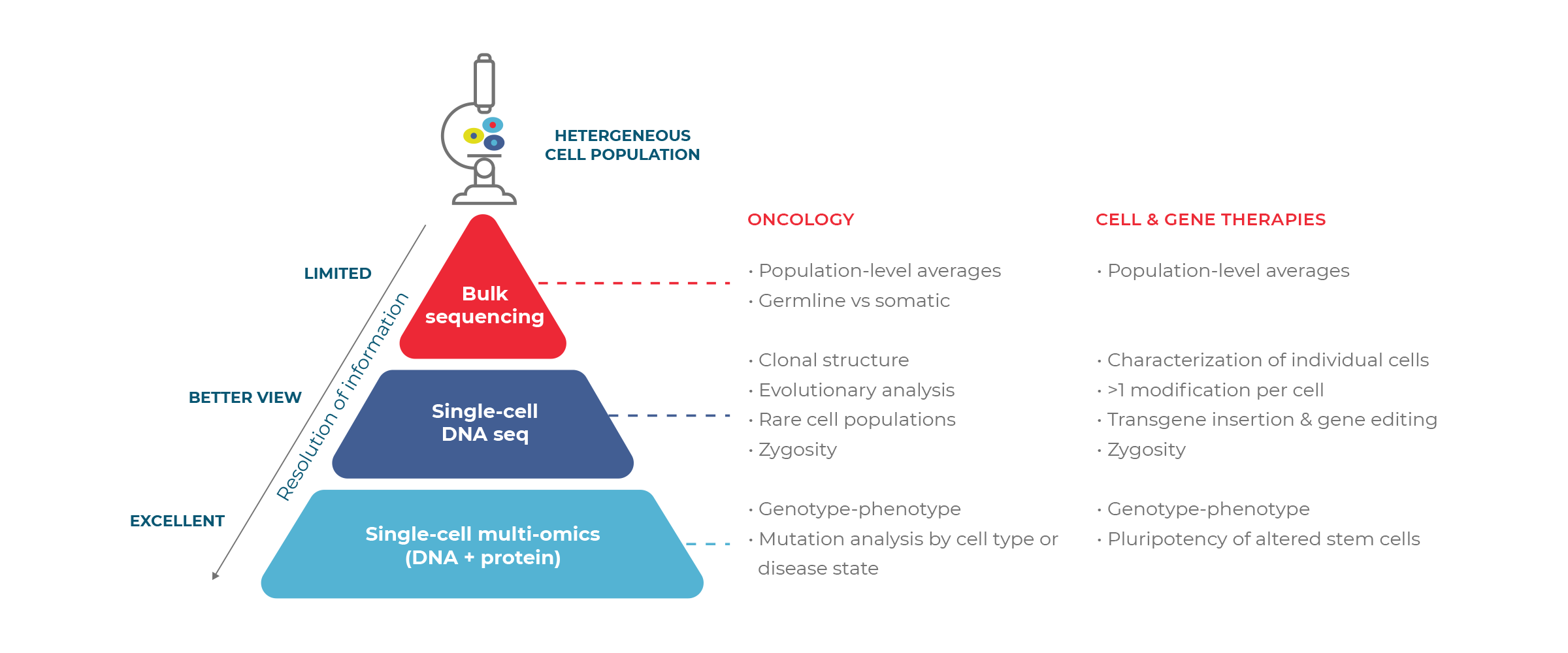
Today, many genotyping workflows rely on bulk assays, which mix DNA across cells and report average readouts. Although appropriate for some questions, these assays do not preserve the information of individual cells, and therefore provide only limited knowledge about biological complexity. Single-cell DNA sequencing, on the other hand, evaluates the genotype of individual cells, and thus provides a richer picture of the sample.
Single-cell sequencing allows for direct measurement of mutations in individual cells by first encapsulating the cells in small droplets and then introducing individual molecular barcodes, enabling researchers to confidently identify which mutations are present in which cells, and no longer need to infer things like zygosity and mutational co-occurrence.
This is especially pertinent for applications where information at the cellular level matters. Some of the most pressing challenges include elucidating the complexity of heterogeneous diseases like cancer, and the underlying mechanisms driving them. Other hurdles lie within the process of pharmacological development, in which each therapeutic candidate must be characterized with sufficient granularity to ensure safety and efficacy.
Single cell Sequencing vs Bulk Sequencing
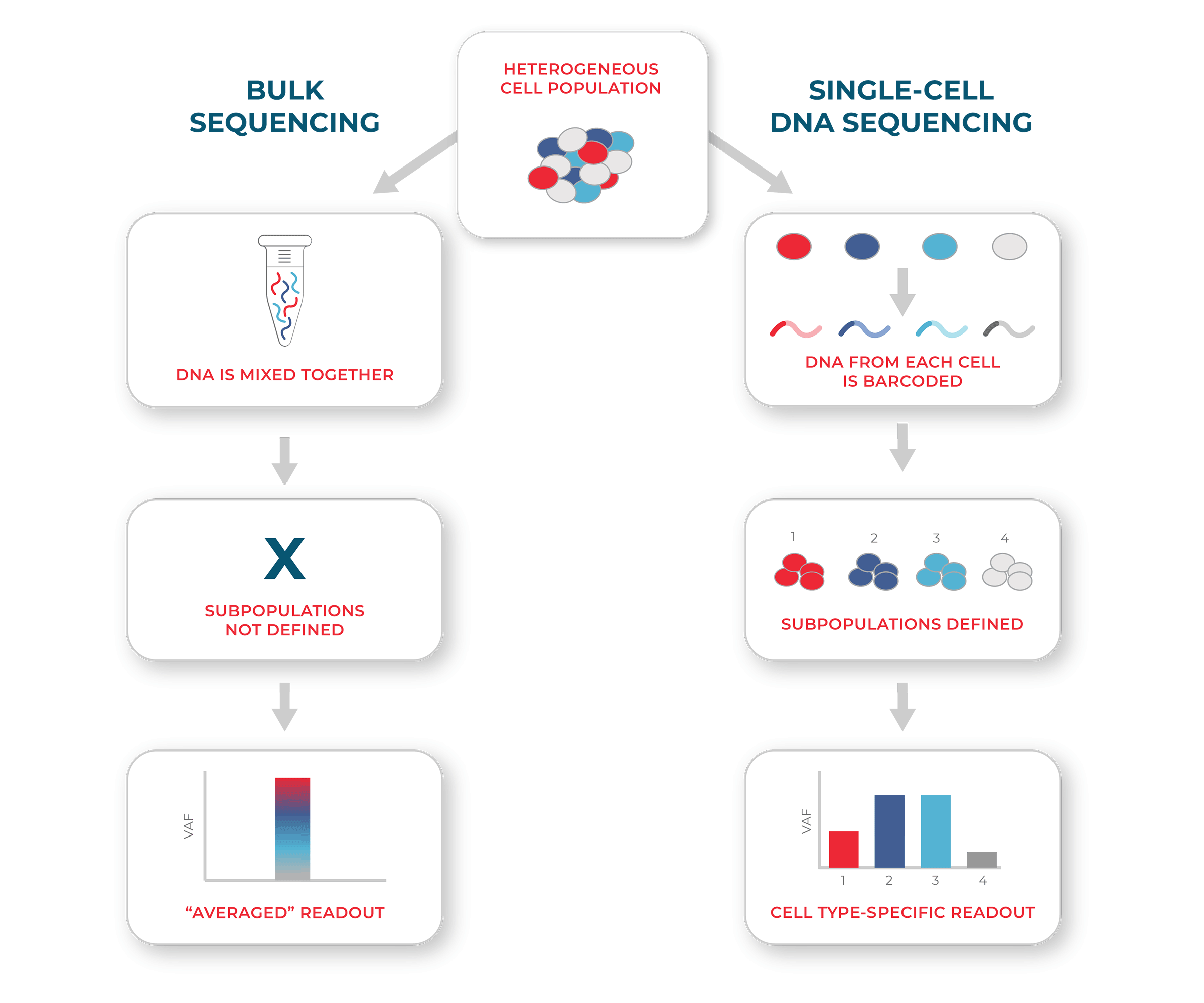
In conventional bulk sequencing (left), DNA is mixed across cells such that genetically distinct subpopulations (e.g. subclones) cannot be identified. Genotypic measurements (e.g. variant allele frequencies, VAF) therefore represent a population average. In single-cell DNA sequencing (right), genetic targets are barcoded in each cell. This allows for genotypic readouts for different subpopulations of cells.