

Myeloid diseases such as AML are highly diverse, with genetic and immunophenotypic changes driving clonal evolution, making response and relapse difficult to predict. Single-cell multiomics helps address this complexity, offering deeper insights into disease progression and treatment response to guide more informed therapeutic decisions.
The complexity of leukemia clonal architecture and the genotypic and immunophenotypic drifts during treatment explain why more than half of patients in remission could relapse. Using single-cell multiomics approaches, researchers can uncover the clonal and subclonal drivers of therapy resistance and relapse, and more precisely characterize patient tumor heterogeneity and evolution.
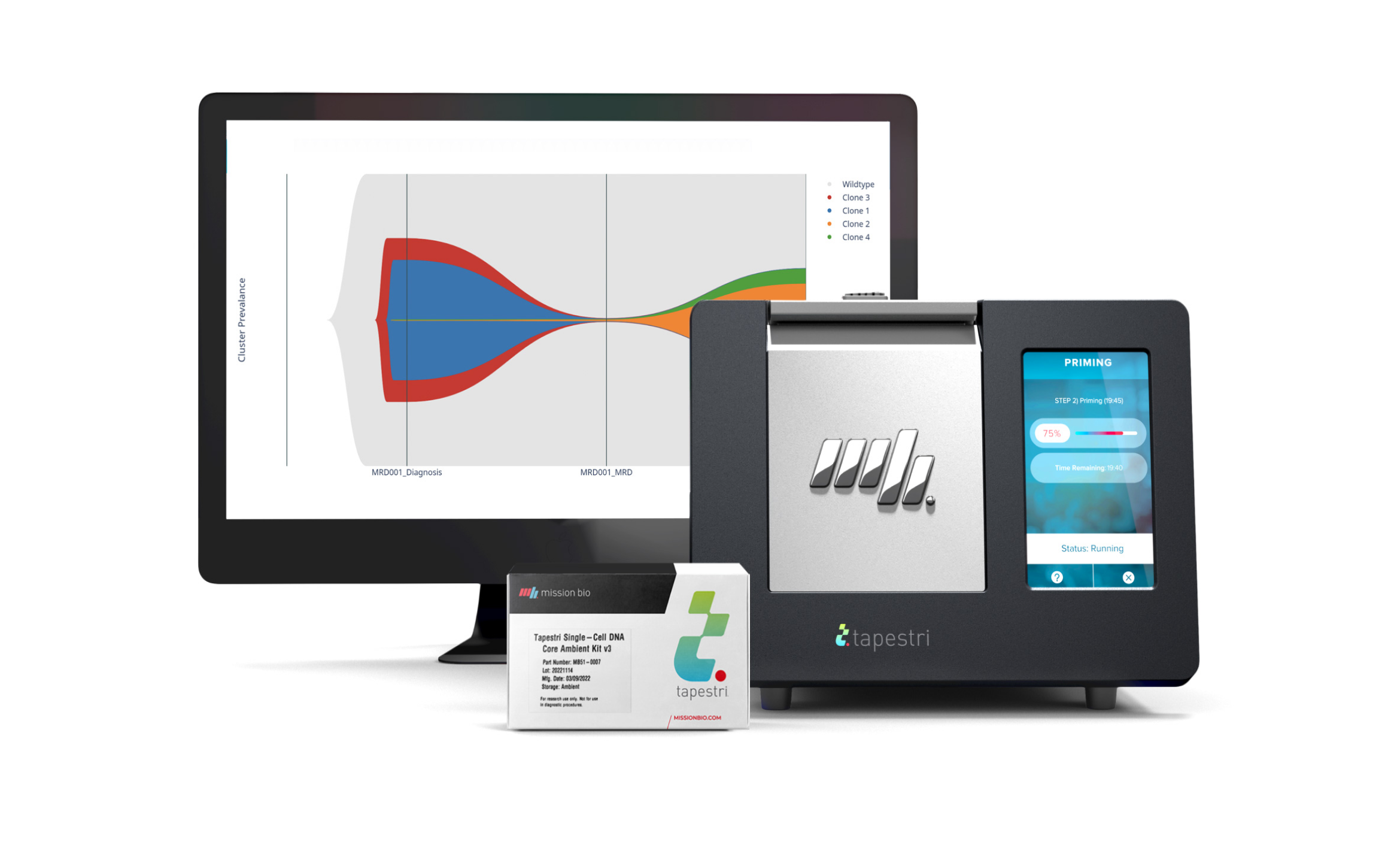
The Tapestri Myeloid Workflow
Panels can be purchased together or separately to suit your research needs.
Panel
Targets Covered
41 hotspot genes based on updated guidelines for acute myeloid leukemia progression, therapy resistance, and relapse.
37 hotspot genes commonly mutated in myeloid malignancies.
31 hotspot genes commonly mutated in myeloid malignancies providing insights into the pathogenesis of myeloid transformation.
17-plex antibody-oligonucleotide conjugate (AOC) panel curated for key biomarkers associated with AML.
P.J.M. (Peter) Valk , PhD, Principal Investigator at Erasmus MC
Independent Mission Bio Platform User
Contact us to begin your evaluation.
For Research Use Only. Not for use in diagnostic procedures.
