

Enhance your research with insightful single-cell bioinformatics and a powerful cohort analysis module optimized for large-scale, multiomic datasets across patient populations and timepoints.
Patient samples are inherently heterogenous, consisting of different clones: wildtype, pathogenic yet therapeutically targetable clones and lastly pathogenic clones. Traditional bulk methods like next-generation sequencing (NGS) or flow cytometry provide population-level snapshots, but they often miss critical co-occurring features that can drive therapeutic resistance.
Single-cell multiomic analysis with Tapestri, enhanced by single-cell bioinformatics, uniquely uncovers hidden pathogenic clones by integrating genotype (DNA) and immunophenotype (surface protein) data at the single-cell level.
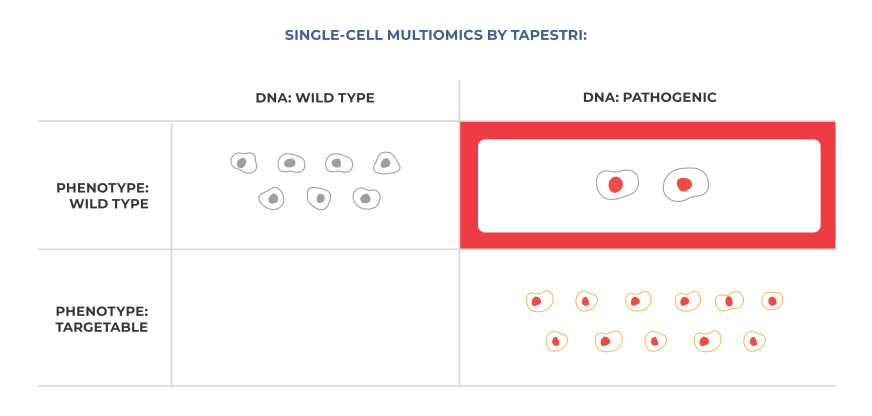
Single-analyte methods like bulk NGS and flow cytometry cannot uncover co-occurring features that may drive therapeutic resistance (top right quadrant). Single-cell multiomic cohort analysis reveals these hidden insights by integrating temporal perspective to genotypic and immunophenotypic data at single-cell resolution.
Key Insights:
Single-cell multiomic analysis enables unparalleled resolution by mapping both genotype and phenotype in individual cells—highlighting clones that would otherwise remain undetected. This integrated view is essential for developing effective therapies and understanding disease progression in complex, heterogeneous samples.
Identify transcriptional and mutational signatures in resistant vs. sensitive patients to uncover pathways driving therapy escape and clonal persistence.
Compare genomic and phenotypic profiles across patient cohorts to distinguish biomarkers and cell states associated with therapeutic response.
Resolve subclonal diversity within and across patients to understand the molecular complexity underlying disease progression and treatment variability.
All cohort analysis begins with high-resolution single-cell sequencing powered by Mission Bio’s Tapestri platform, which captures DNA and protein-level data from each individual cell.
The cohort analysis pipeline, powered by advanced single-cell bioinformatics, combines patient sample and time points from h5 files generated from Mission Bio’s Tapestri platform to generate a heatmap across multiple attributes.
Attributes such as clonal fraction, VDJ clonotypes, prognostic structural variants and counts, as well as focal CNV, mutations and prognostic protein markers can be visualized and trends can be compared across samples, demonstrating clonal dynamics that impact disease progression.
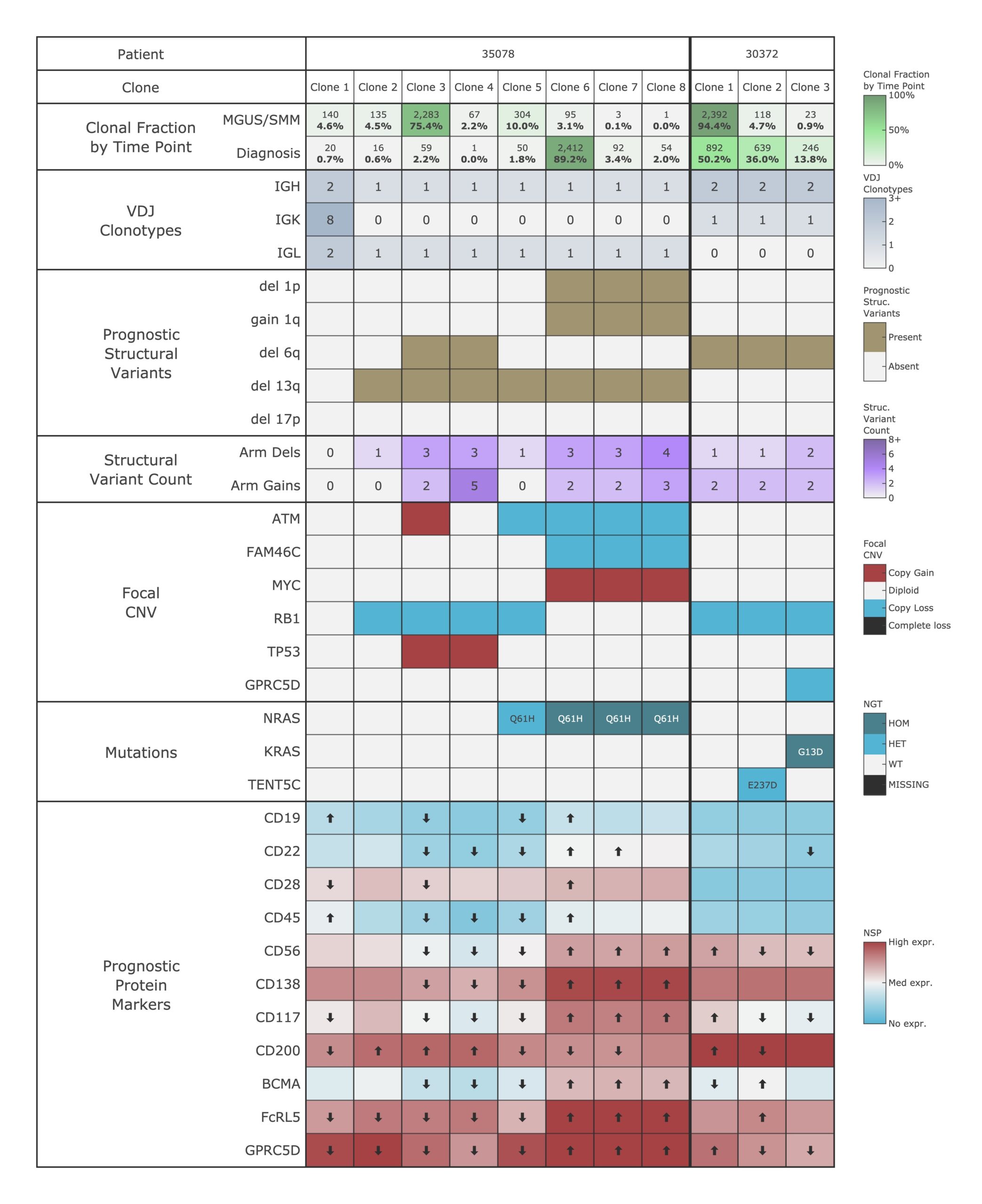
This figure illustrates the clonal landscape of two patients with precursor plasma cell neoplasms (MGUS/SMM), as characterized using the Cohort Analysis pipeline. Each row represents a layer of multi-omic data resolved at the single-cell level, and each column represents a genetically distinct clone identified within the sample.
This multi-dimensional view enables deep characterization of disease heterogeneity and clonal evolution, offering insights into progression risk, therapeutic target expression (e.g., BCMA), and potential for relapse. The Cohort analysis Pipeline on Mosaic is uniquely suited to resolve these complex relationships, empowering translational research and precision medicine in hematologic malignancies.
This phylogenetic tree was generated using the Cohort Analysis pipeline to visualize the clonal evolution and mutational landscape in a patient at Timepoint-1, diagnosed with Monoclonal Gammopathy of Undetermined Significance (MGUS)
Learn more about how Cohort Analysis is powered by Mosaic.
The diagram illustrates the clonal structure of hematopoietic cells, highlighting the genetic heterogeneity present at the single-cell level. Each node represents a genetically distinct clone, annotated with its specific genomic alterations and the proportion of total cells it comprises.
This clonal map reveals a branched evolution pattern, where a wildtype progenitor gives rise to progressively complex subclonal populations. The high frequency of Clone 3 suggests strong clonal dominance, while subclones with NRAS mutations may indicate early oncogenic events contributing to disease progression. These findings underscore the value of single-cell DNA analysis for early detection and monitoring of clonal dynamics in precursor conditions like MGUS.
This clonal phylogenetic tree illustrates the evolutionary relationships and genomic alterations present in a patient sample at Timepoint-2 (Diagnosis), as resolved using the Mission Bio Tapestri® single-cell DNA sequencing platform.
Each branch represents a distinct genetically defined clone, annotated with:
Tapestri-powered single-cell phylogenetic reconstruction enables high-resolution insight into tumor heterogeneity and clonal fitness, supporting therapeutic risk stratification and molecular monitoring strategies.

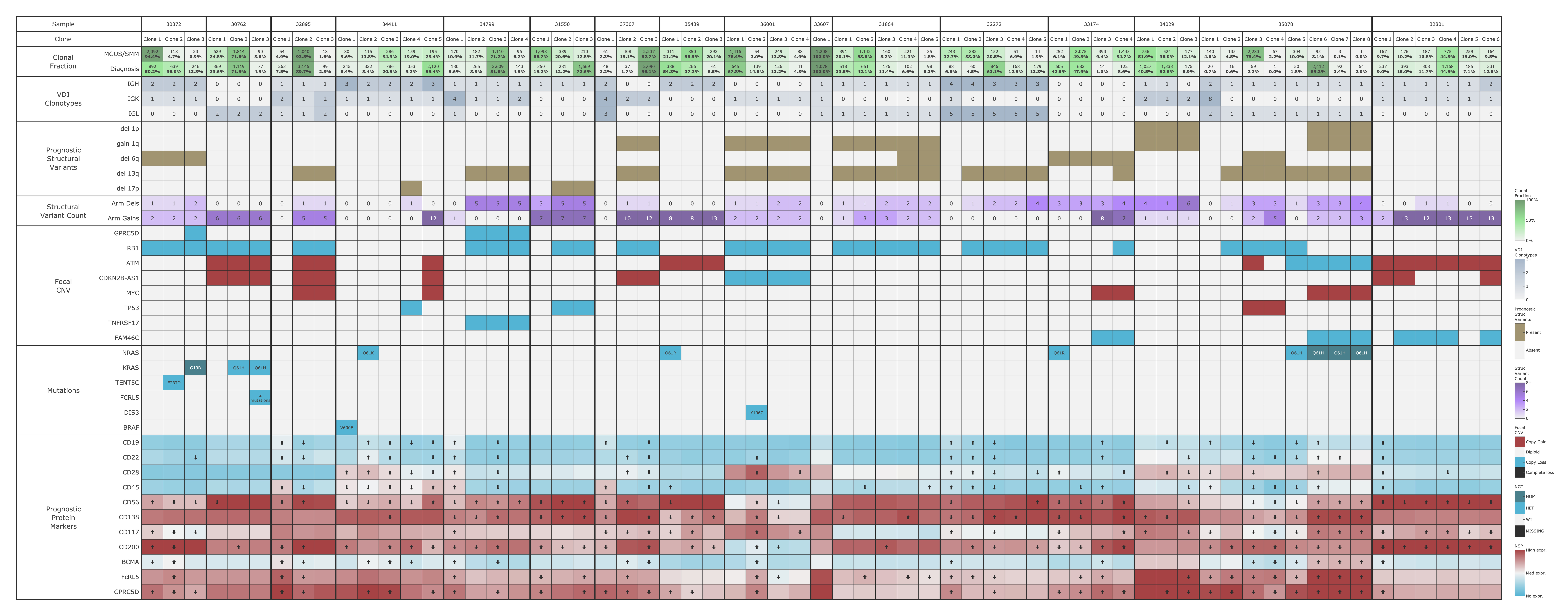
Figure: Single-Cell Multi-Omic Profiling Across 16 Patients with Plasma Cell Neoplasms
This figure showcases data from 16 patients with monoclonal gammopathies, including MGUS and smoldering multiple myeloma (SMM), analyzed using the Mission Bio Tapestri® Mosaic Cohort Analysis pipeline. Each patient’s sample is deconvoluted into genetically distinct clones, with integrated insights across DNA, protein, and structural features.
Key data layers include:
This cohort analysis highlights the ability of Mission Bio’s single-cell multi-omics and single-cell bioinformatics to resolve disease heterogeneity, map clonal evolution, and assess clinically actionable features, supporting precision oncology efforts in plasma cell malignancies.
*The content provided herein may relate to products that have not been fully validated by Mission Bio and is subject to change without notice.
For Research Use Only. Not for use in diagnostic procedures.
Figure: Single-Cell Multi-Omic Profiling Across 16 Patients with Plasma Cell Neoplasms



