

Understanding the genetic variability of multiple myeloma at the single-cell level is crucial for clinical researchers to break the treatment resistance and relapse cycle. Curated based on input from global key opinion leaders in multiple myeloma and peer-reviewed studies, this panel targets the most common mutations in driver and therapy-resistance genes associated with multiple myeloma, as well as indels, mutational zygosity, focal copy number aberrations, and BCR IgH/IgK/IgL gene rearrangements.
LEARN MOREAdvance your understanding of the genetic heterogeneity underpinning multiple myeloma by targeting hotspot mutations in driver & resistance genes, focal copy number aberrations, and V(D)J clonotype with 339 amplicons for single-cell DNA sequencing. This panel has been verified in a wet lab to meet performance specifications.
DOWNLOAD DATASHEET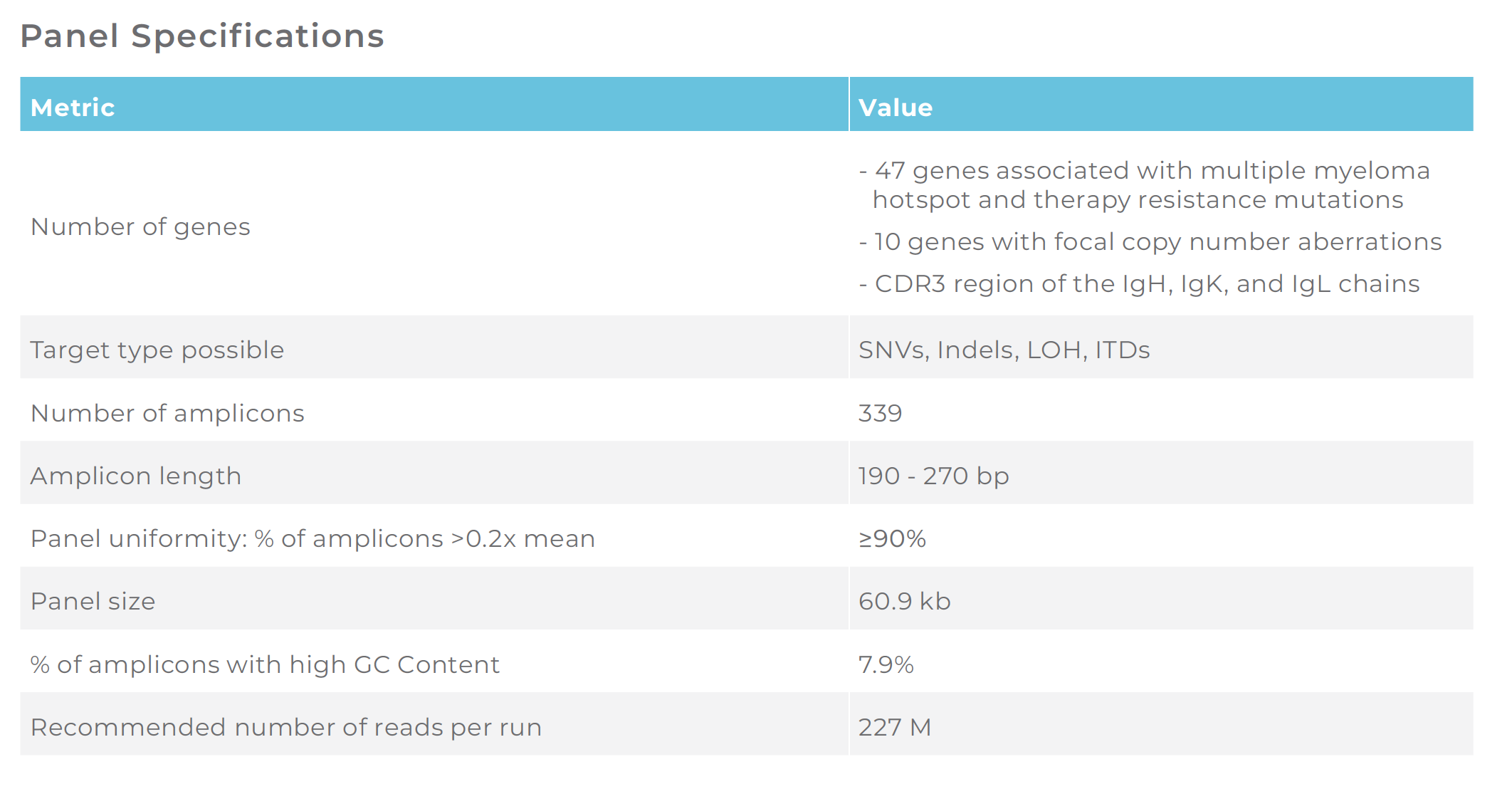

Use Tapestri Designer to add or remove content from existing catalog panels and make this panel your own with content that is relevant to your research. Or start from scratch and upload your human (hg19) or mouse (mm10) targets using gene names, genomic coordinates, or SNV IDs. Target anywhere in the whole human or mouse genome and advance your research with targeted single-cell DNA sequencing.
DESIGN PANEL