How do four alphabet letters (nucleotides) give rise to the vast diversity of biological life? April 25th is DNA Day, which celebrates key milestones in the search for the answer to this question about the molecular code of life.
At Mission Bio, we strive to actively push the field of genomics forward by helping researchers analyze DNA at the single-cell level. Using our Tapestri Platform, researchers are performing comprehensive multi-omics analyses and detangling the complexities of diseases, like cancer, at an unprecedented level of detail. Although the day specifically commemorates the discovery of DNA’s double helical structure in 1953 and the completion of the Human Genome Project almost exactly fifty years later, modern molecular biology wouldn’t be where it is without the countless contributions made by other scientists over the past century and a half.
Join us for a stroll through the history of genetics to learn more about some of the landmark discoveries that have transformed our thinking about biology and shaped our modern world.
At Mission Bio, DNA is in our DNA!
Enter to win our DNA Day 2021 t-shirt

A Stroll Through the History of Genetics
1866 — The Fundamentals of Inheritance. Our promenade starts in 1866 with the discovery of the basic principles of genetics by Gregor Mendel. Often referred to as the “Father of Genetics,” Mendel laid the foundation for understanding of heredity with his fundamental laws of inheritance.
Speculating on the basis of genetic inheritance, Mendel proposed that pea plants pass on a physical material, which he termed “elementen”, to their offspring. Two years later, Charles Darwin postulated a similar mechanism of inheritance. He hypothesized that particles of inheritance, called “gemmules,” are shed by cells and congregate in the gonads (to be passed onto offspring).
Activity: Interpret pedigrees to understand how inherited diseases are passed on.

Mendel discovered the rules of inheritance by studying pea plants
1869 — The Discovery of DNA. The year the Swiss physiological chemist Friedrich Miescher discovered DNA in used, pus-coated surgical bandages — of all places! Calling DNA “nuclein”, Miescher wrote, “It seems probable to me that a whole family of such slightly varying phosphorous-containing substances will appear, as a group of nucleins, equivalent to proteins.” Miescher’s named his discovery “nuclein” because it came from the nuclei of cells, but today we call it deoxyribonucleic acid, or DNA.
Activity: Extract DNA from strawberries!

Friedrich Miescher
Image: Science Museum Group (CC BY 4.0)
1944 — DNA as a Unit of Heredity. Seventy-five years after the discovery of DNA — Oswald Avery and his colleagues identified DNA as hereditary material. Together with Colin MacLeod and Maclyn McCarty, Avery showed that DNA was the ‘transforming principle’, a term coined by Frederick Griffith in 1928 that referred to a substance transferred between bacteria (a process known as transformation). Griffith showed that this substance from a killed strain of bacteria could confer biological properties in a living strain, but did not know what the substance was. The work by Avery and his colleagues confirmed that the molecular carrier of hereditary information was DNA and not protein (a popular belief at the time).
Activity: Explore the inside of a bacteria cell with this interactive simulation.
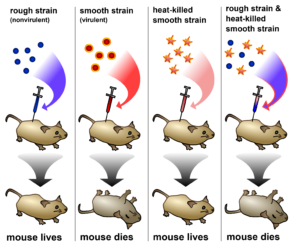
Griffith’s experiment demonstrated his “transforming principle”. The nonvirulent strain of pneumococcal bacteria became lethal when mixed with a heat-killed preparation of the virulent strain, suggesting that the live bacteria were able to acquire properties from the dead bacteria.
Image: Madeleine Price Ball (CC0/public domain)
1953 — Constructing the Double Helix. The race to discover the structure of DNA heated up in the 1950s. Although Linus Pauling was the first to propose a structure (a triple helix), it was disproved soon after when James Watson and Francis Crick proposed their double helix model on April 25, 1953. Watson and Crick arrived at the double helix by building physical models from cardboard and metal scraps.
This seminal discovery would have not been made without help from Rosalind Franklin, whose X-ray crystallography was critical to determining the 3D helical structure of DNA. Although Watson and Crick and another scientist (Maurice Wilkins) received the Nobel Prize for their contributions, Rosalind was not recognized at the time for her important work.

A metal plate representing a nucleotide in Watson and Crick’s DNA model
Image: Mrjohncummings (CC BY-SA 2.0)

X-Ray Crystallography of DNA by Rosalind Franklin
Image: Ryan Somma (CC BY-SA 2.0)
1960 — Decoding Codons. Just over a decade later, the American scientist Marshall Nirenberg decoded the words (codons) that make up the language of life. The first codon (the one encoding phenylalanine, or UUU) was discovered in 1960 by spiking ground-up bacterial cells with a synthetic RNA molecule containing only uracil in what’s known as the “poly-U experiment”. The remaining codons would be discovered in the five years that followed.
Activity: Discover the triplet code that encodes all 20 amino acids.

Marshall Nirenberg with a personalized license plate displaying the first codon he discovered (UUU for phenylalanine)
Image: NIH (public domain)
1973 — The Birth of Genetic Engineering. The field of genetic engineering was born in 1973 when Stanley Cohen, Herbert Boyer, and colleagues published the first paper describing recombinant DNA technology. In the paper, they showed that biologically functional DNA fragments could be joined together in plasmids that retained the functional properties of each of the joined DNA fragments.
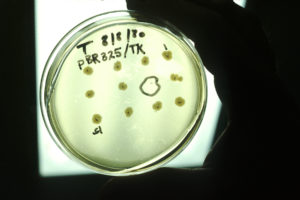
An experiment using recombinant DNA technology. Here, recombinant DNA was used to express the thymidine kinase (TK) gene from the herpes simplex virus in bacteria, rendering them susceptible to the tetracycline antibiotic
Image: National Cancer Institute (public domain)
1977 — Sequencing the Code of Life. Frederick Sanger developed the “rapid DNA sequencing” technique known today as Sanger sequencing. The original method uses chain-terminating nucleotides and gel electrophoresis to define the sequence of nucleotides in a given DNA strand. Refined versions of this method would be eventually used to sequence the human genome as part of the Human Genome Project. The Sanger method continues to be used for small-scale DNA sequencing today.
Activity: Watch a video to explore how Sanger sequencing works.
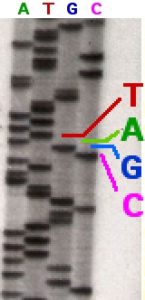
A radioactively labeled sequencing gel depicting the Sanger method
Image: John Schmidt (CC BY-SA 3.0)
1987 — Making Transgenic Organisms. It wasn’t long from the birth of genetic engineering that the first transgenic animals were created. The first transgenic mice were generated in 1987 by injecting DNA into mouse embryo cells, as described in landmark papers published in Nature and Science. A year later, transgenic fruit flies were described in another Science paper. The development of these transgenic animals led to the realization that slight modifications to gene sequences could be used to study gene function.
These demonstrations opened the door for genetically modifying other organisms, as speculated in a 1981 article on the discoveries: “Leaving aside the controversial question of gene therapy in humans, it seems certain that the genetic manipulation of agriculturally important animals will quickly follow…”
Activity: Understand the science of transgenic technology by watching this video.

A series of transgenic fruit flies containing mutations in genes responsible for eye and cuticle color
Image: public domain
1990 — The Human Genome Project. With the commercialization of Sanger sequencing technology by Applied Biosystems in 1986, the stage was set to sequence the human genome. The Human Genome Project began in 1990. Its goal was to sequence the entire euchromatic human genome in just 15 years. As depicted in the project’s logo, the potential of this project for wide-reaching impact in a variety of fields was not lost on scientists.

Logo of the Human Genome Project
Image: U.S. Department of Energy, Human Genome Project (public domain)
1996 — First Cloned Mammal. Using an adult mammary gland cell, Dolly the sheep was the first mammal to be cloned. The achievement not only showed that a cell from one part of the body could be used to recreate a whole individual, but it also demonstrated that a differentiated, adult cell could revert to an embryonic, totipotent state.
Activity: Learn about how shotgun sequencing enabled the sequencing of the human genome.

Dolly the sheep is the first cloned mammal
Image: Toni Barros (CC BY-SA 2.0)
2002 — Completion of the Mouse Genome. The full impact of the Human Genome Project would have been lessened without the concurrent sequencing of other genomes, namely those of model organisms like the laboratory mouse. Recognizing this, the NIH called for a working draft of the laboratory mouse genome in 1998 and set a deadline of 2003 to achieve it. The project finished early in 2002. The feat represented the first completely sequenced mammalian genome.
Sequencing the genome of the laboratory mouse genome was a landmark event
The first completed mouse genome could not have come at a better time, as The Human Genome Project “finished” ahead of schedule only a year later. The project was declared complete in April 2003, with 99% of the euchromatic human genome sequenced at 99.9% accuracy. (The euchromatic human genome accounts for about 90% of the total human genome and excludes telomeres and centromeres.)
2005 — Next Generation-Sequencing (NGS). With the mouse and human genomes sequenced (as well as several other organisms), the scientific community recognized that faster and more efficient technologies would be needed to sequence a much larger number of organisms. Spurred by the continued industrialization of sequencing technologies, biotech companies including Roche, Solarex, and Life Sciences developed and commercialized the next generation of sequencers by 2005. These next-generation sequencing technologies would exponentially accelerate the pace of genomics research in the years to come.
Activity: Watch a video about how sequencing technologies have improved cancer diagnosis and treatment.
A flow cell used for next-generation sequencing
Image: Bainscou (CC BY 3.0)
2012- CRISPR Gene Editing Tool. Our tour ends in 2012, the year Jennifer Doudna and Emmanuelle Carpentier repurposed the bacterial CRISPR-Cas9 system as a programmable gene-editing device. The scientific community at large was stunned by the discovery, immediately realizing the dramatic impact it would have on life science research and precision medicine. Since 2012, the CRISPR toolbox has been expanded to include gene activation and inhibition technologies as well as more refined editing tools like base editors.
Activity: Explore an interactive module to learn how CRISPR works.
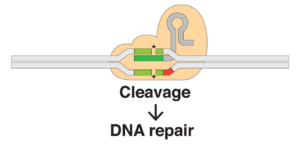
The CRISPR-Cas9 gene-editing tool
Image: Mariuswalter (CC BY-SA 4.0)
DNA and Our Modern World
It’s been less than 200 years since DNA was discovered, and yet its presence can be felt throughout today’s society. Modern forensics, agriculture, ecology, medicine — you name it! — would look much different without the genetic and genomic advances that have defined our current way of life. We can now improve crop yields, combat genetic diseases, and rapidly diagnose and develop treatments against novel pathogens, among other amazing feats.
Even so, challenges remain in the development of precision medicines for more complex diseases, like cancer. Understanding DNA is key to deciphering and hopefully curing these devastating illnesses. At Mission Bio, we strive to enable researchers to better understand the genetic architecture of tumors through the use of single-cell DNA sequencing and multi-omics.
Overall, the exponential clip of genomics research, including the continual advancement of DNA technologies, promises exciting times ahead — for cancer therapeutics and more!
Still thinking about that DNA Day t-shirt? Go ahead, enter to win!











