

Myeloid diseases such as AML are highly diverse, with genetic and immunophenotypic changes driving clonal evolution, making response and relapse difficult to predict. Single-cell multiomics helps address this complexity, offering deeper insights into disease progression and treatment response to guide more informed therapeutic decisions.
The complexity of leukemia clonal architecture and the genotypic and immunophenotypic drifts during treatment explain why more than half of patients in remission could relapse. Using single-cell multiomics approaches, researchers can ucover the clonal and subclonal drivers of therapy resistance and relapse, and more precisely characterize patient tumor heterogeneity and evolution

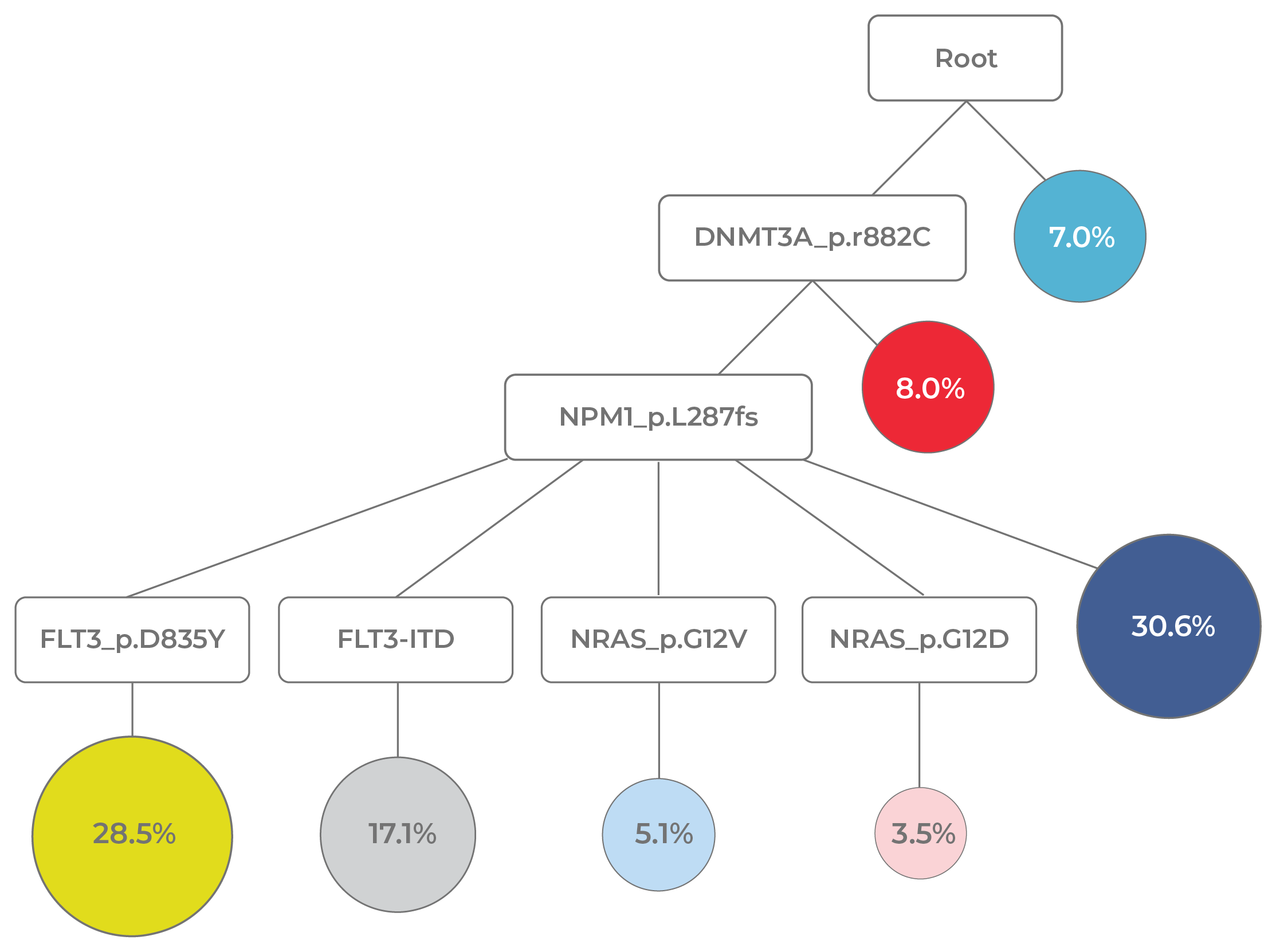
The heterogeneity and dynamism of AML clonal architecture pose a challenge for MRD monitoring. Yet, MRD assessment is crucial for prognosis, as MRD positivity post-remission indicates a higher relapse risk. Conventional single-analyte methods like flow cytometry and PCR are unable to detect the immunophenotypic or genotypic changes arising during treatment. While error-corrected NGS offers highly sensitive detection of residual cells and simultaneous evaluation of many genes, it cannot differentiate true AML from precursor clones, leading to false-positive results. Even when combining these assays to achieve clinical utility, integrating data from different modalities remains challenging.
P.J.M. (Peter) Valk, PhD, Principal Investigator at Erasmus MC
Independent Mission Bio Platform User
Contact us to begin your evaluation.
For Research Use Only. Not for use in diagnostic procedures.
