In a recent webinar, Adam Sciambi, CTO, Mission Bio, presented new data from a multiple myeloma study specifically looking at paired samples from pre-diagnosis time points such as monoclonal gammopathy of undetermined significance (MGUS) and mid-stage smoldering multiple myeloma (SMM), and diagnostic time point (active MM). He discussed, using a single-cell multiomics approach, how we are able to assign a diverse set of molecular attributes to subclones within that sample, understand why the disease progressed, and also how therapy might be guided in the future.
What is Multiple Myeloma?
Fig. 1. Illustration of the MGUS, SMM, and Diagnostic stages of MM.What is Myeloma?, digital illustration, MSKCC, accessed 29 May 2024, https://www.mskcc.org/cancer-care/types/multiple-myeloma
Multiple myeloma (MM) is a genetically diverse disease hallmarked by the clonal expansion of plasma cells. This clonal outgrowth makes MM incredibly difficult to treat acutely—patients often relapse after treatment and require multiple treatments, with failures often attributed to the clonal heterogeneity of the disease. There are often a variety of clones with different variances associated with them, as well as variants associated with resistance to treatment that could also cause treatment failure.
Multiple Myeloma is Genetically Diverse
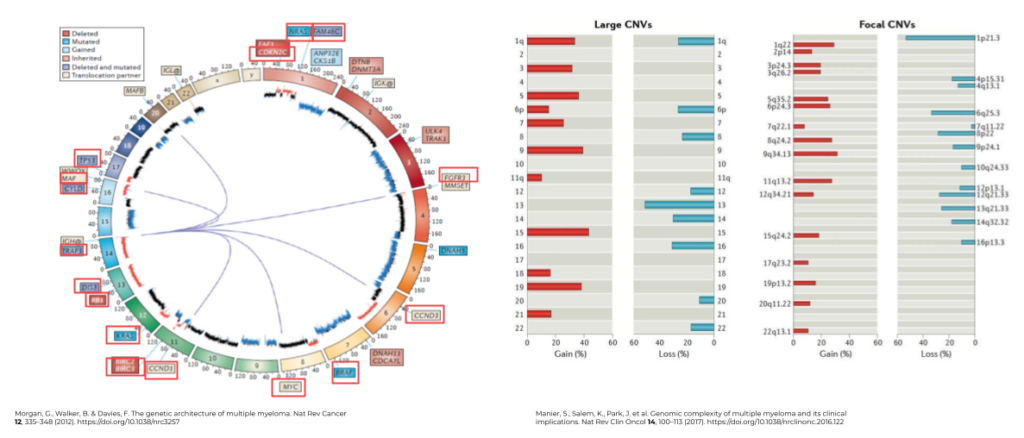
Fig. 2. The genetic architecture and genomic complexity of Multiple Myeloma.
One facet that makes MM so difficult to treat is that the disease inherently has many different molecular drivers and indicators. MM is a genetically complex malignancy comprised of immunoglobulin translocations, focal and genome-wide CNVs, complex genomic variations in myeloma-associated mutations, and epigenetic modifications. Figure 2 illustrates the multitude of point mutations and driving genes (Fig 2, left) associated with MM, as well as structural variants (Fig 2, right), such as chromosomes associated with copy gain (hyperdiploidy), copy loss, and focal copy numbers, that can be associated with more advanced disease.
Emerging Precision Myeloma Therapies
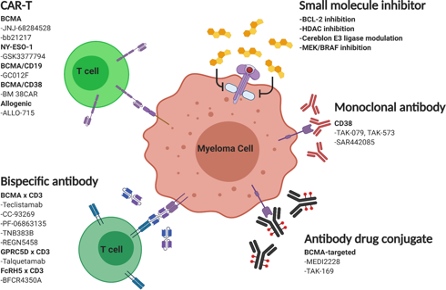
Fig. 3. Emerging therapies for relapse/refractory Multiple Myeloma. Su, C.T., Ye, J.C. Emerging therapies for relapsed/refractory multiple myeloma: CAR-T and beyond. J Hematol Oncol 14, 115 (2021). https://doi.org/10.1186/s13045-021-01109-y
There are many molecular signatures driving MM, and over the past several years, targeting tumor surface proteins has rapidly emerged as one of the most exciting frontiers for treatment options. Figure 3 highlights some of these markers and various therapies, such as CAR-T, bispecific monoclonal antibodies, antibody-drug conjugates (ACs), and inhibitors that can help treat the disease.
Multiple Myeloma has a High Relapse Rate
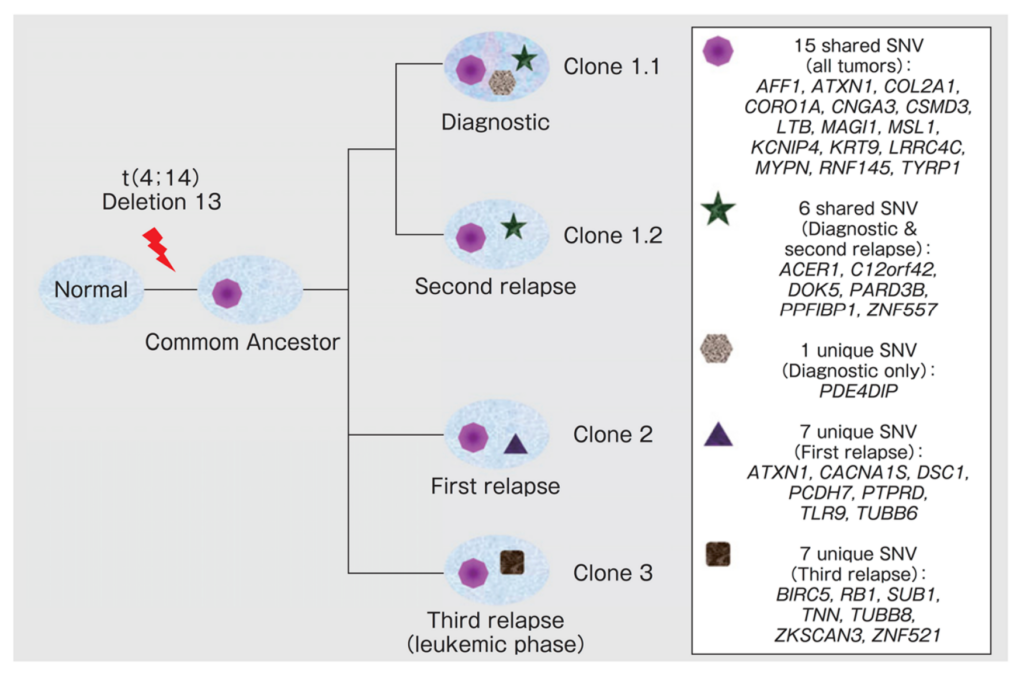
Fig. 4. Illustration of clonal hierarchy and expansion in MM. J Egan, et al. Blood 120, 1060 (2012).
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3412329/
Despite several diagnostic techniques available and advancements in immunotherapies for MM, the disease carries high relapse & mortality rates, suggesting that these approaches remain inadequate in eradicating the disease. These techniques could benefit from a more precise method to pinpoint the clonal heterogeneity involved in disease evolution and therapeutic resistance, from the earliest stage of MGUS to SMM and eventual full-blown MM.
Figure 4 illustrates bulk sequencing data from a particular MM patient and how, through several rounds of treatment, different clones are brought to the fore because of treatment resistance. The Diagnostic time point shows the various initial clones present, followed by the clones present after treatment. Upon the first relapse, Clone 2 becomes dominant, and with different subsequent treatments, Clone 1.2 becomes more dominant. Upon the third relapse, Clone 3 appears.
This diversal architecture makes for challenging therapy selection because one needs to be able to treat many different clones with potentially mutually exclusive therapeutic targets. This data highlights the necessity of truly understanding clonal heterogeneity to better treat disease.
Molecular Signatures of Multiple Myeloma
By looking at a more complete list of potential molecular signatures for MM, we can better track and understand the disease.
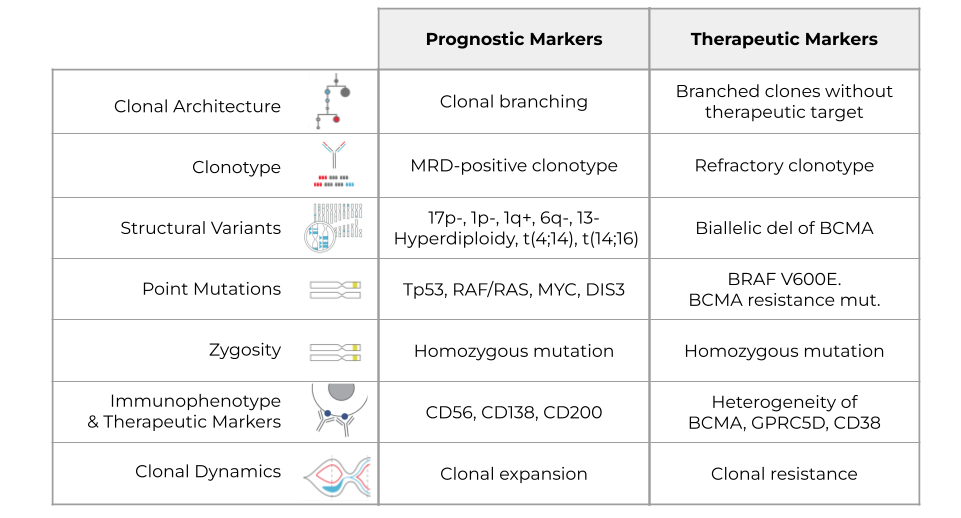
Clonal Architecture
The first molecular signature to look at is clonal architecture. Assessing clonal architecture enables you to understand which clones are present in the patient and how they are spread. Understanding clonal architecture lets you see if evolution is linear or branched. Branched clonal architecture may be an indicator of poor prognosis, whereas linear clonal architecture may indicate a better prognosis. If the clonal architecture is very branched, that means that there are many different competing clones, and any one of them can cause the next relapse.
With clonal architecture, it would be therapeutically insightful to know whether your target, which you may have gleaned from flow cytometry or NGS, is present or not in all clones. That could tell you which clone might cause relapse after treatment.
Clonotype
The next signature is clonotype. The BCR VDJ signature helps you understand clonality and is often one of the earlier mutations you’ll see, which helps you distinguish healthy, wild-type samples from mutants. If the patient is in remission, a prognostic marker might be an MRD-positive clonotype, a rare clonotype detected at diagnosis and now reappeared. As for therapeutic guidance, if you have a refractory clonotype, that most likely means your therapy is not working.
Structural Variants
Multiple myeloma is a disease of genomic instability, so understanding structural variants helps you identify bad-acting clones, even if they don’t carry driver mutations. There are a lot of prognostic and therapeutic targets associated with structural and copy number variants. For example, there’s a loss of 17p- associated with TP53, in addition to a loss of 1p, a gain of 1q, and so on. Hyperdiploidy translocations are prognostic indicators as well. Additionally, if you have a biallelic deletion of BCMA, that indicates that your BCMA therapy might fail.
Point Mutations
Point mutations are important drivers of MM and can be targeted for therapy. Some examples of prognostic markers include TP53, RAF/RAS, MYC, DIS3 and many others. There are also several potential therapeutic targets like BRAF and V600E. BCMA resistance mutation indicates that your BCMA therapy might not work.
Zygosity
Because a homozygous mutant is often more deleterious than a heterozygous mutant, zygosity plays an important role as an indicator. Zygosity may be informative for treatment purposes as well – a homozygous mutation might actually mean that it responds better to therapy targeting that pathway because of the reliance of that particular cell on this mutation to propagate.
Immunophenotype & Therapeutic Markers
Understanding immunophenotype and therapeutic markers is important as well. These indicators provide prognostic insight into the disease and also give you potential therapeutic targets. For example, CD56, CD138, or CD200 all indicate a poor prognosis. If you have heterogeneity in BCMA, GPRC5D, or CD38 (these are all therapeutic targets), your therapy might not work as well, assuming that all of the mutants express these targets.
Clonal Dynamics
Lastly, you want to understand clonal dynamics or how the disease changes and evolves. Clearly, clonal expansion gives you a poor prognosis. A therapeutic indicator of this could be that if you see clones that aren’t responding (shrinking), you have therapy resistance. Many clones will respond, but some might not, and that would be an indicator that your therapy is not working.Current
Assays for Measuring MM Attributes
When treating MM, it’s essential to understand the therapeutic and prognostic markers listed here. But how do current technologies stack up for measuring these attributes? Here, we highlight some of the conventional technologies that are used to investigate these signatures.
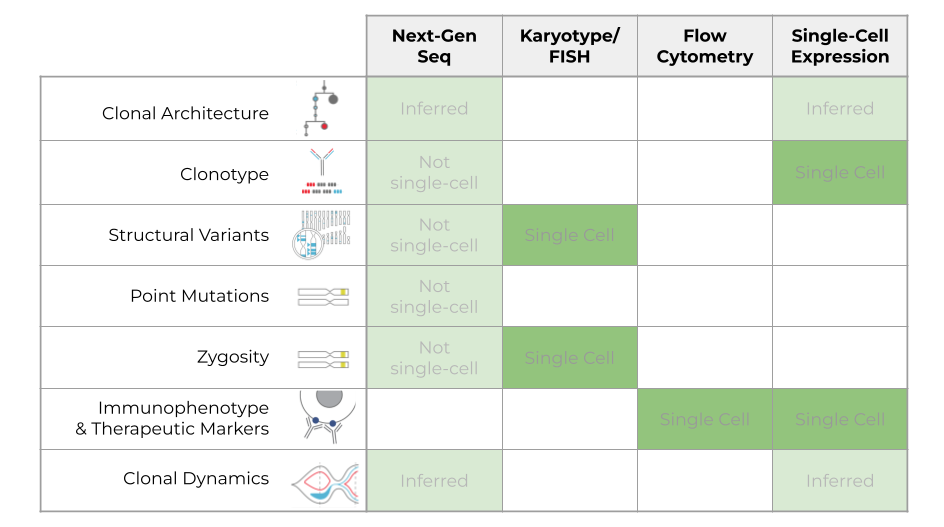
One of the most versatile technologies is Next-Generation Sequencing, or NGS, which can measure many of these attributes but can only infer clonal architecture and clonal dynamics. You can also measure clonotype, structural variants, point mutations, and zygosity with NGS, but not at the single-cell resolution, so you can’t tell how these populations are grouped together or whether these features are mutually exclusive or co-occurring.
Karyotyping/FISH is another important way to analyze multiple samples. It’s a single-cell technology but is limited in what it can look at regarding structural variants and zygosity. This technology is widely used for diagnosing samples but is limited in what attributes it can analyze.
Flow cytometry is similarly very powerful, but again, this technology is limited to measuring protein attributes like immunophenotype, whereas karyotype and FISH are more informative of structural or copy number variants.
Lastly, single-cell gene expression analysis is a widely used research tool. With this technology, you can only infer clonal architecture and clonal dynamics. Additionally, you can look at clonotype and phenotype, but there’s no commercially available platform that can look at all of these together.
Gain Insights into MM Clonal Heterogeneity and Evolution with Tapestri
Here, we take a look at these same attributes, but now with a demonstration of what the data output looks like from the Tapestri Platform.
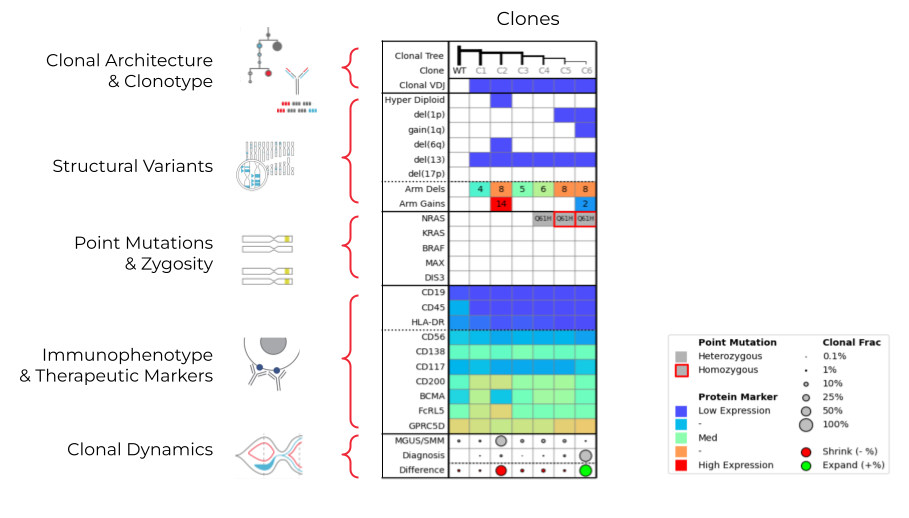
The example data table above shows how each row corresponds to a subset of these attributes on the left. In the first row, you can see the branching clonal architecture, as well as the V(D)J clonotype. You can see the structural variants—hyperdiploidy, del(1p), and del(17p), as well as arm-level deletions and gains. The point mutations row shows whether a mutation is homozygous or heterozygous. You can also see the immunophenotypes present—CD19, CD45 are more typical of healthy cells, while CD56, CD138, BCMA, and FcRL5 are all prognostic and therapeutic markers. Finally, the clonal dynamics row shows the population size of clones at different time points.
Each column represents a clone measured against all of these analytes. With bulk sequencing, this information is averaged together, but with single-cell multiomics enabled with the Tapestri Platform, we can see these different genotypes and phenotypes within the patient at the subclonal level. Let’s take a deeper look to understand how these different attributes contribute to the dynamics of these clones.
In the MGUS sample, Clone 2 dominated, and Clone 6 was only detected at 0.7%. By the time of diagnosis, Clone 6 had expanded to 94%. Clone 2 uniquely has hyperdiploidy, del(6q), and many arm-level gains. Clone 2 also seems to have a lower expression of BCMA, but a higher expression of FcRL5. You can see from the MGUS to the diagnostic time point that Clone 2 appears to be shrinking.
On the contrary, Clone 6 expands over time, potentially due to the combination of bi-allelic NRAS mutation (from HET to HOM), del(1p), and gain(1q). In addition to identifying features associated with clonal expansion, Tapestri identified putative therapeutic targets (e.g. BCMA, RAS, CD200), which could be potentially used to guide treatment in the future.
The Tapestri platform helps unveil this inherent complexity in MM samples by looking at all these attributes simultaneously and holistically and pinpoint very rare clonal populations that will expand long before it happens. In this example, it identified rare clones at the SMM/MGUS time points that expand by the diagnostic time point. Bulk NGS lacks the ability to measure rare clones and clonal populations.
Mission Bio has developed the Tapestri Single-cell Multiple Myeloma Multiomics Solution, which allows researchers to gain crucial insights into the clonal heterogeneity and evolution underlying myeloma progression, therapy response, and relapse often overlooked by traditional bulk methods.
For Research Use Only. Not for use in diagnostic procedures.










