Tumors represent intricate ecological systems in which diverse clonal subpopulations interact and co-evolve within the context of their microenvironmental niches. The process of clonal evolution, spanning from the initial neoplastic lesions to the terminal metastatic disease state, is a continuous phenomenon that molds cancer progression in response to both cell-intrinsic and extrinsic factors impinging upon the tumor. Although the traditional metastatic cascade model remains widely embraced, it fails to fully encapsulate the emerging paradigms that govern the intricate dynamics of tumor evolution and the extensive heterogeneity that arises during this process.
Methods
In our investigation of clonal evolution in pancreatic cancer, we utilized an innovative in vivo platform termed Clonal Replica Tumors (CRTs). This platform facilitates the generation of animal cohorts harboring human tumors that share an identical clonal barcode composition. The CRTs were derived from multiple patient-derived xenograft (PDX) models exhibiting high biological clonal diversity, as validated through single-cell DNA sequencing (scDNA-seq) analyses. These clonally diverse CRTs originated from PDX models displaying distinct organotropism towards the liver and lungs, allowing us to trace the dynamics of pancreatic ductal adenocarcinoma (PDAC) clones during tumor expansion and metastatic dissemination over a 3-month timeframe. By employing this approach, we examined the metastatic process as an inherent aspect of the complex pancreatic tumor ecosystem, elucidating the continuous clonal evolutionary trajectory beyond single endpoint assessments.
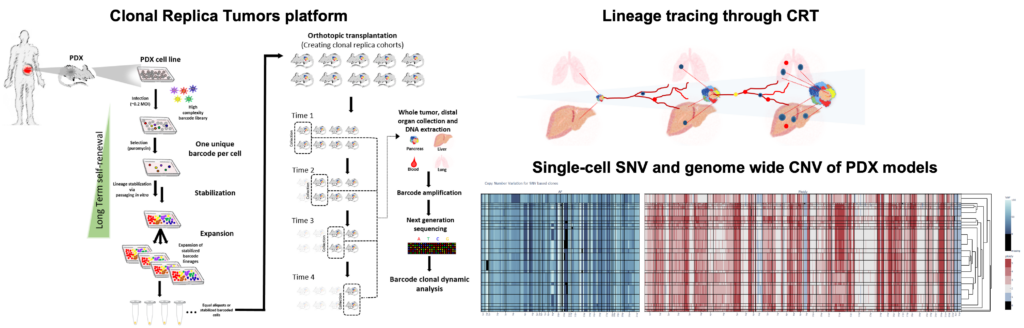
Results
Defying conventional expectations, the majority of clonal lineages within primary tumors did not exhibit a straightforward pattern of continuous fitness gain or loss. Instead, these lineages displayed a phenomenon termed alternating clonal dominance (ACD), wherein clones continuously fluctuated in their relative representation, leading to a constant reshuffling of the tumor’s clonal architecture in the absence of external perturbations. Remarkably, a substantial fraction of lineages contributing to primary tumor growth disseminated to secondary sites during tumor progression, even in non-metastatic tumors, strongly implying that tumor dissemination is a generalized, non-specific behavior. Nonetheless, only a select few lineages possessed the capability to form definitive metastatic lesions, while many underwent transient expansion before disappearing, a phenomenon we termed abortive colonization.
Our findings revealed that cell-intrinsic factors, such as self-renewal capacity and clonal fitness, exhibited a strong correlation with the ability to form metastatic lesions. Furthermore, through the isolation and characterization of isogenic subclones with pro- and non-metastatic phenotypes, we identified a transcriptomic signature that enabled the identification of metastatic cells within human tumors and predicted patient survival outcomes.
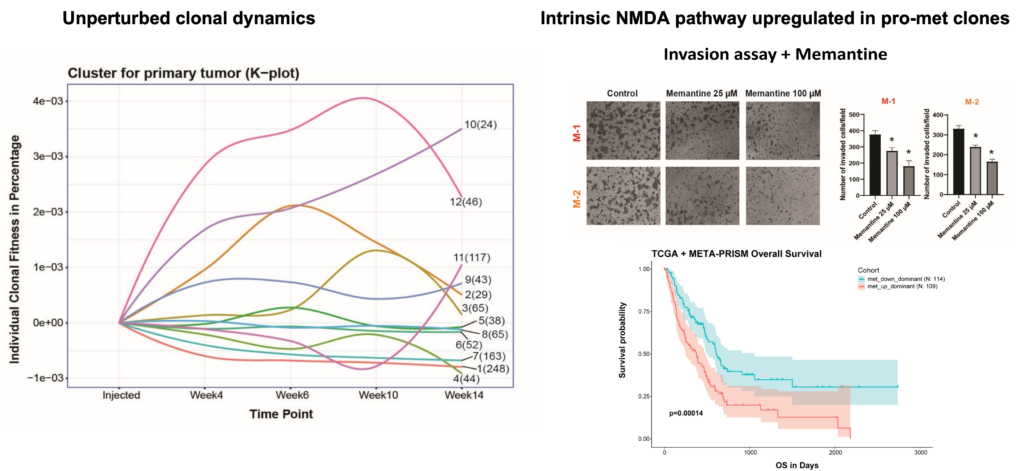
Conclusions
The Clonal Replica Tumor (CRT) platform represents a significant technological leap forward in the modeling of human disease, shedding light on an unprecedented level of subclonal intricacy inherent to tumors undergoing unperturbed expansion and dissemination processes. By capturing the intricate subclonal dynamics within these unmanipulated tumor ecosystems, CRTs have unveiled a remarkable degree of complexity that was previously unappreciated when studying tumor evolution and metastatic spread.










