Research Background
Acute myeloid leukemia (AML) is a complex disease with a diverse genetic landscape, leading to treatment challenges and frequent relapses. Measurable residual disease (MRD) testing has become a standard in the management of AML, utilized to detect remaining leukemia cells post-treatment and guide risk assessment and treatment decisions for better outcomes. However, monitoring MRD in AML is challenging due to its evolving nature.
In this study, the team led by Ross Levine, MD, and Wenbin Xiao, MD, PhD, at MSK demonstrates the capabilities of the Tapestri® Single-Cell MRD AML Multiomic Assay to identify relapse-related mutations missed by bulk with a limit of detection of 0.01%, and characterize the clonal architecture and distinguish pre-leukemic from leukemic subpopulations with combined single-cell DNA sequencing and immunophenotyping.
How the Tapestri Platform helped
A major limitation of using bulk next-generation sequencing (NGS) data to interpret MRD results and outcomes is its inability to distinguish MRD clones from preleukemic or clonal hematopoiesis (CH) clones. This distinction is crucial for accurately predicting the risk of relapse.
Single-cell MRD (scMRD) assay can more accurately characterize MRD by discerning distinct clones within a patient while simultaneously identifying genotype-specific changes in protein expression patterns over the course of disease and treatment. This highly precise clonal identification can potentially inform targeted therapeutic approaches.
In this study, the authors analyzed 30 AML MRD samples using the Tapestri Platform. By combining mutational and immunopheno-typic assessment in a single assay, they were able to characterize relapse-driving MRD clones at unprecedented resolution and identify clinically relevant clones that bulk was unable to detect (Figure 1).
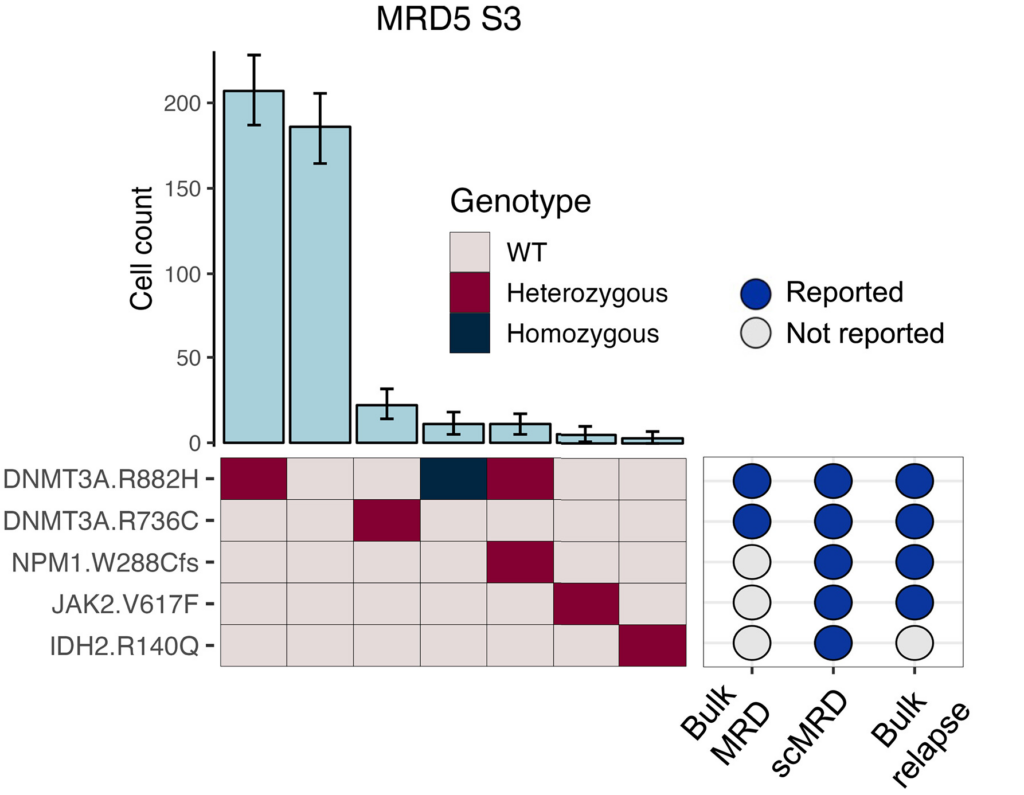
Figure 1: Presence of different clones within a patient, including preleukemic DNMT3A, bystander JAK2, and relapse-related DNMT3A/NPM1 clones. Bulk NGS failed to detect JAK2 and NPM1 mutations at the remission time point but identified them at the subsequent relapse time point.
Additionally, the authors noted the differential immunophenotypic states between CH/preleukemic and leukemic clones within and between patients (Figure 2). CH clones exhibited similar immuno-phenotypes to wild-type, while leukemic clones consistently showed aberrant immunophenotypes. For example, single-mutant TP53 clones with higher CD71 expression and DNMT3A/NPM1 co-mutated cells expressed CD11b and CD16 granulocytic/ monocytic markers.
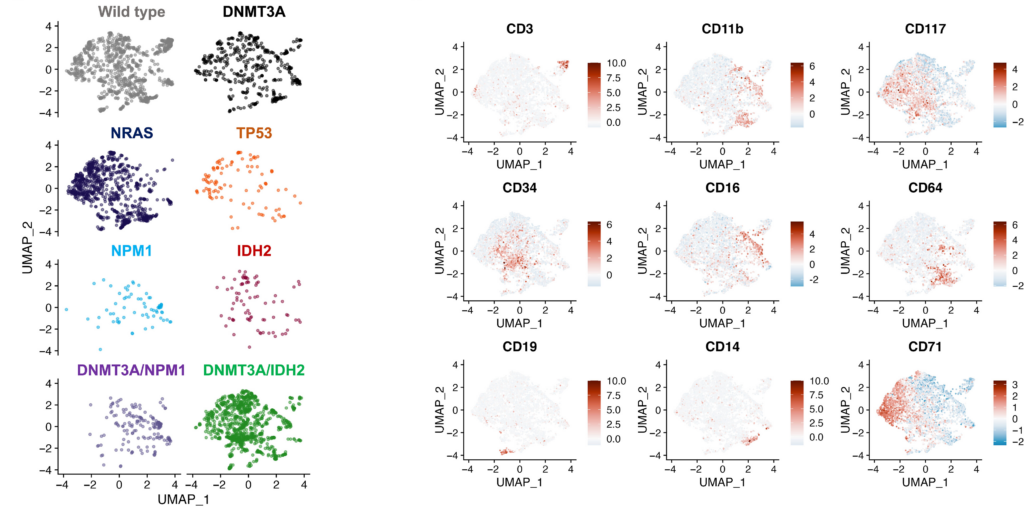
Figure 2: UMAP analysis identifying the differential immunophenotypic states of single-mutant versus compound-mutant clones.
These data highlight that integrated genomic and immunopheno-typic analysis at the MRD time point has the potential to discern the nature and behavior of the residual cells left after treatment. This understanding is essential for making informed decisions regarding treatment strategies and monitoring the risk of relapse in individuals with AML.
Tapestri Single-Cell MRD Multiomics AML Assay
The Tapestri Single-Cell MRD Multiomics AML Assay simultaneously conducts and integrates the genotypic and immunophenotypic assessment of AML MRD across thousands of individual cells, providing clonal insight in tandem with immunophenotype from rare residual disease cells. These high-resolution, integrated molecular profiles bring unprecedented clarity to the complex biology of AML MRD, powering actionable insights.
The Tapestri Single-Cell MRD Multiomics AML Assay enables researchers to:
-
Reveal clonal architecture and uncover the order of acquisition of mutations
-
Track clonal dynamics over time through the course of the disease and treatment to identify potential theraputic targets and therapy-resistant clones
-
Unambiguously distinguish true MRD from Preleukemic or precursor clones with a limit of detection of 0.01%
The Tapestri Single-Cell MRD Multiomics AML Assay is now available. To learn more, check out some of the resources below:










