Understanding how single-cells behave in a heterogeneous cell population, whether within a tumor microenvironment or a therapeutic product, is critical for advancing both oncology and cell & gene therapy (CGT) programs. With regulatory agencies providing more stringent guideline requirements to demonstrate the safety and efficacy of these therapies, traditional bulk analysis methods may not provide enough data clarity needed to be approved by the agencies in the coming years. By moving beyond bulk- or population-level data averaging and getting granular with analysis on the single-cell level, researchers can gain a more profound understanding of cellular identity and function, leading to better, safer, and effective therapies that save lives.
To address the inherent limitations of bulk analysis, researchers are increasingly employing single-cell analysis methods to obtain a comprehensive understanding of disease biology, identify relevant therapeutic targets and biomarkers for drug development, and establish assays capable of evaluating therapeutic persistence and safety following administration. Importantly, these methods also facilitate patient stratification for clinical trials by identifying molecular subpopulations most likely to respond to treatment, improving therapy trial design, enrollment strategies, and the evaluation of the therapeutic safety.
By partnering with companies like Mission Bio who offer custom, fit-for-purpose Pharma Assay Development (PAD) services, researchers can develop end-to-end assays for their therapy, from early discovery to assay transfer for clinical applications and commercialization, that are tailored to the specific needs of their oncology and cell & gene therapy programs.
Already want to build a custom, fit-for-purpose assay for your therapy? Let’s chat.
Advantages of single-cell assays vs bulk assay
The fundamental challenge in oncology and cell & gene therapy is that the human body is a mosaic of cellular diversity that is particularly evident in how diseases are developed; between genetic mutations, gene expression dysfunctions, and irregular protein accumulations. Tumors, for example, are not homogenous cell masses but are dynamic ecosystems consisting of malignant cancer cells, diverse array of immune cells, and stromal cells like cancer-associated fibroblasts (CAFs). This cellular heterogeneity fuels cancer progression, metastasis, and the development of therapeutic resistance. Similarly, in the early development of engineered cell therapies, the final drug product, the cell, is rarely uniform. Genetically modified cells can exhibit significant cell-to-cell variability in their on-target edits, off-target events, genome integrity, and resulting therapeutic function.
Traditional methods such as bulk DNA sequencing and bulk RNA sequencing provide an averaged snapshot of a cell population. In these methods, nucleic acids from thousands of millions of different cells are extracted, pooled, and sequenced together. The resulting data reflects the overall mutational profile or gene expression of the entire sample, but it fundamentally loses all information about individual cells within that population. This averaging can obscure critical details. For example, a rare, drug resistant cancer subclone or low-frequency population of highly potent therapeutic cells would be diluted and go undetected in a bulk sequencing analysis. This bulk sequencing analysis is not a technical inconvenience, it is a major flaw in our approach to understanding the complexities of biological diseases, because it leads to a mischaracterization of disease by treating a heterogeneous population as a single entity.
For biotech and pharmaceutical companies seeking to develop successful therapies, the inability to resolve cellular heterogeneity across genetic, transcriptional, and proteomic layers introduces significant risks into both discovery and clinical translation. Partnering with a single-cell multi-omics analysis provider that offers Pharma Assay Development (PAD) services directly addresses these challenges by enabling simultaneous measurements of DNA, RNA, and protein within the same cell in a sample population. This single-cell multi-omics data resolution provides critical information into how genomic alterations translate into gene expression and ultimately influencing the phenotype; all information that is essential for identifying the biomolecular drivers of the disease to the right therapeutic responses. To be more direct, these Pharma Assay Development services are customizable and fit-for-purpose based on the needs of the researcher and the therapeutic program.
Oncology: Bulk analysis methods miss clonal diversity and transcriptional shifts
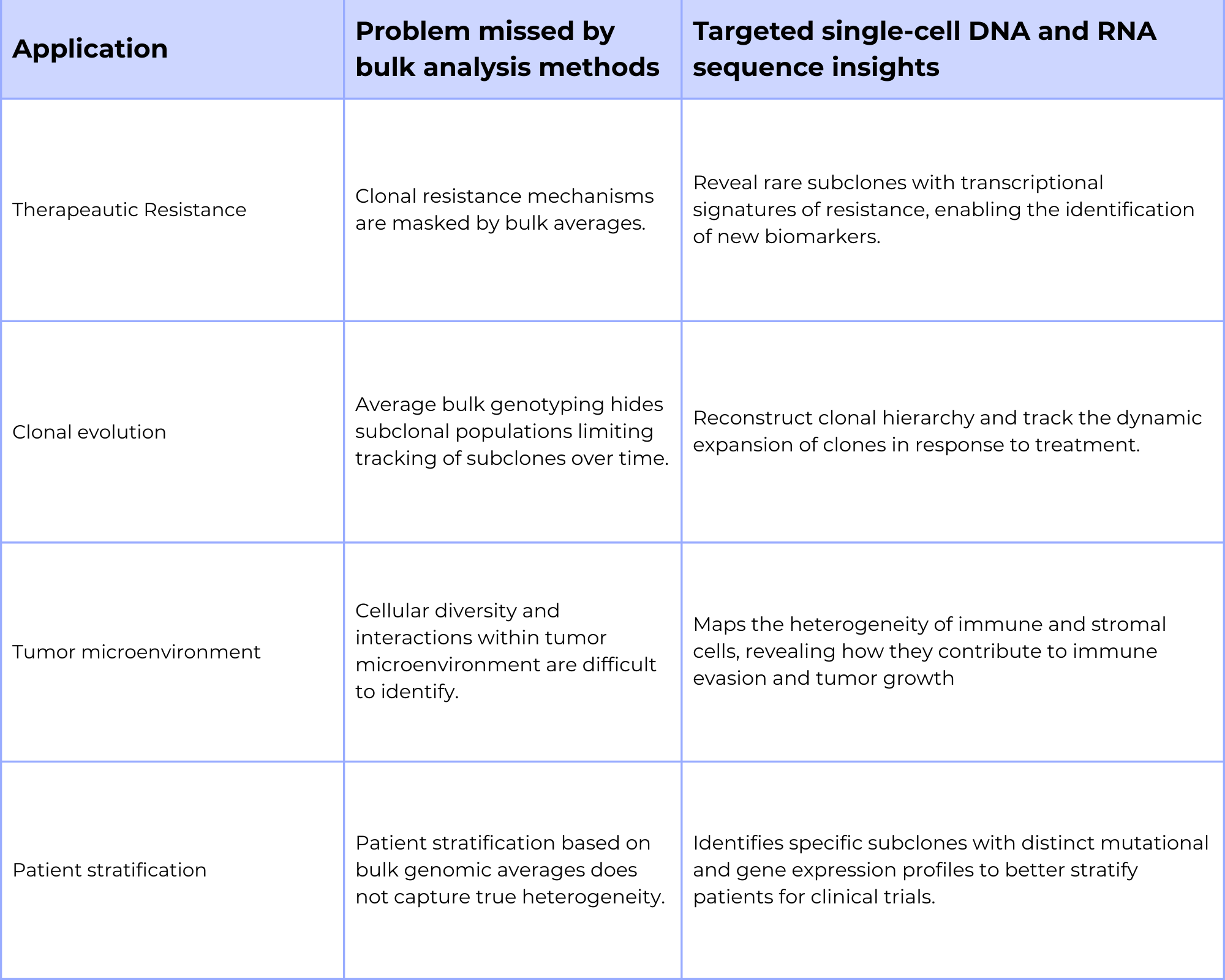
Through the accumulation of mutations and selection, tumors generate significant genetic and phenotypic heterogeneity, which often leads to the acquisition of therapeutic resistance. A major challenge for oncology research is understanding how this clonal evolution occurs and identifying the specific mechanism of resistance (MoR) before they lead to clinical relapse.
Detecting rare subclones with transcriptional features of resistance
Targeted single-cell DNA analysis on platforms such as Mission Bio’s Tapestri platform allows for the precise measurement of single-nucleotide variants (SNVs), copy number variations (CNVs), and structural characteristics like zygosity and co-occurences of mutations in thousands of individual cells. This high resolution genotypic data enables researchers to reconstruct clonal hierarchy and track how tumors evolve over time in response to treatment. This is particularly useful for identifying rare, drug-resistant clones that may pre-exist in tumors or emerge under therapeutic pressures. Such rare subclones, which would be masked by bulk DNA sequencing methods, can be the cause of relapse.
Taking this a step further, single-cell analysis that incorporates DNA sequencing and gene expression profiling enables researchers to dissect mechanisms of resistance (MoR) at high resolution. For example, it can identify rare drug resistant cancer stem cells and reveal whether resistance arises from upregulation of a specific gene or through the activity of a co-activator within a subclone that drives a functional resistance phenotype. By linking genetic alterations, transcriptional changes, and phenotypic responses at the clonal level, single-cell approaches uncover the causal relationships that underlie therapeutic failure. Under selective pressure, clonal evolution favors the expansion of resistant subpopulations, which can be precisely detected and characterized through single-cell targeted DNA sequencing and gene expression profiling and other single-cell multi-omics methods.
Identify genotype to phenotype discordance linked to relapse
While DNA mutations often initiate disease progression, their downstream effects are mediated through gene expression that drive cellular phenotypes. However, not all mutations translate into functional phenotypes, a discordance that can only be resolved through single-cell multi-omics.
Mission Bio’s PAD services can help address these challenges by simultaneously profiling genomic mutations, targeted RNA expression, and phenotype of the same cell, enabling researchers to directly connect genotype to its phenotype. This capability is critical for dissecting the complexity of the tumor microenvironment, where genetically distinct cancer subclones interact with diverse immune and stromal populations differently. By linking these mutations to gene expression and transcriptional states, such as immune activation or stem resistance mechanisms, researchers can uncover what specific mutations drive phenotypes associated with relapse and therapeutic resistance.
Genetic mutation → gene expression → phenotypic response → relapse or resistance
Bulk analysis methods also obscure both pathogenic drivers and actionable biomarkers. Single-cell multi-omics empowers researchers to understand the genotype-to-phenotype discordance enabling them to stratify patients more precisely, anticipate, replace, and design better tailored therapies for the patient that directly target the functional biomolecular drivers of disease.
Stratify patients based on expression signatures tied to drug response
Beyond elucidating mechanism of resistance (MoR), targeted single-cell multi-omics is a powerful tool for patient stratification. By simultaneously capturing mutations and gene expression within individual cells of a sample, this approach distinguishes clinically relevant subclones that would otherwise be obscured in bulk sequencing analysis methods. Such high resolution profiling enables the identification of patient subgroups with unique genetic drivers, transcriptional signatures, or pathway activations that influence therapeutic responses. This level of single-cell insights allows researchers to match patients to the therapies most likely to be effective, refine clinical trial enrollment strategies, and reduce variability in trial outcomes. Ultimately, integrating gene mutation and gene expression data with single-cell granularity not only increases the probability of trial success but also advances the broader goal of delivering the right therapy to the right patient at the right time.
Table 1: Advantages of targeted single-cell DNA and RNA analysis in oncology
Cell & gene therapy (CGT): Connecting genome editing outcomes to downstream functional impact
Engineered cell therapies, including CAR-T and gene edited stem cells, are heterogenous by nature, reflecting both intrinsic cellular diversity and the complexity of their production. This puts a strong emphasis on the safety and efficacy of these cell therapies from development to clinical trials, from small scale studies to scaling up for commercial manufacturing. This variability can significantly impact the therapy’s safety, potency, and persistence. Targeted single-cell analysis methods provide the high-resolution needed to fully characterize these therapies by linking genetic attributes to the resulting gene expression and cellular function.
How to link gene edits to gene expression and functional impact for cell therapies
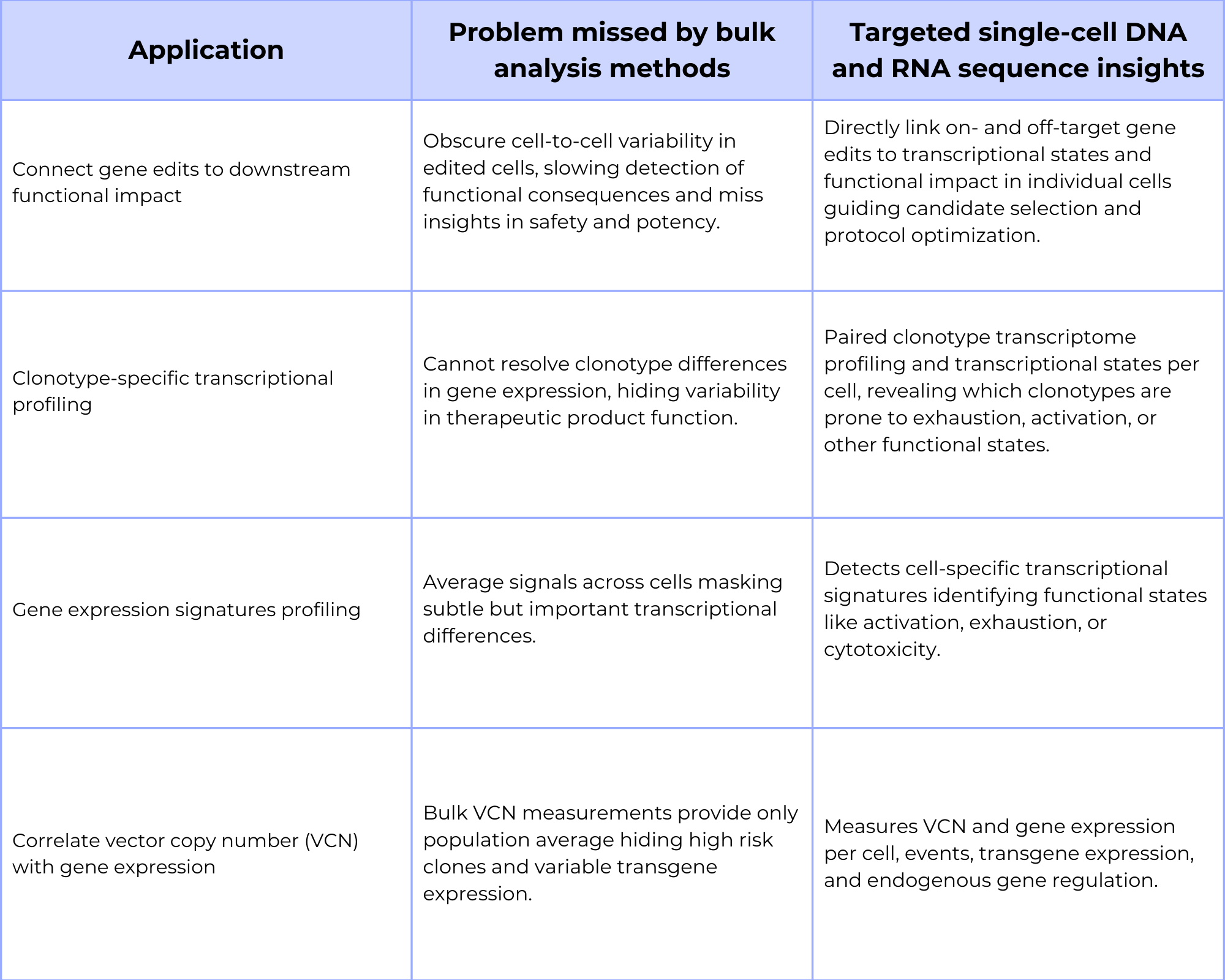
Cell and gene therapies that use gene editing tools like CRISPR, it is crucial to understand not only if the gene edit was successful, but also what impact it has on the gene expression and transcriptional state. Conventional techniques and analytical methods used during cell and gene therapy development, such as clonal expansion and bulk assays, are slow and can obscure cell-to-cell variability due to averaging, often requiring weeks before meaningful data comes is collected. Single-cell multi-omics methods overcome these limitations by simultaneously detecting gene edits and their gene expression or transcriptional states enabling direct linkage between specific on- and off-target edits, gene expression, and the downstream functional impact all in the same cell within the cell therapy. This can help guide lead candidate selection and refine gene editing protocols to maximize the workflow efficiency during therapy development.
Clonotype-specific transcriptional profiling to understand variability in drug product function
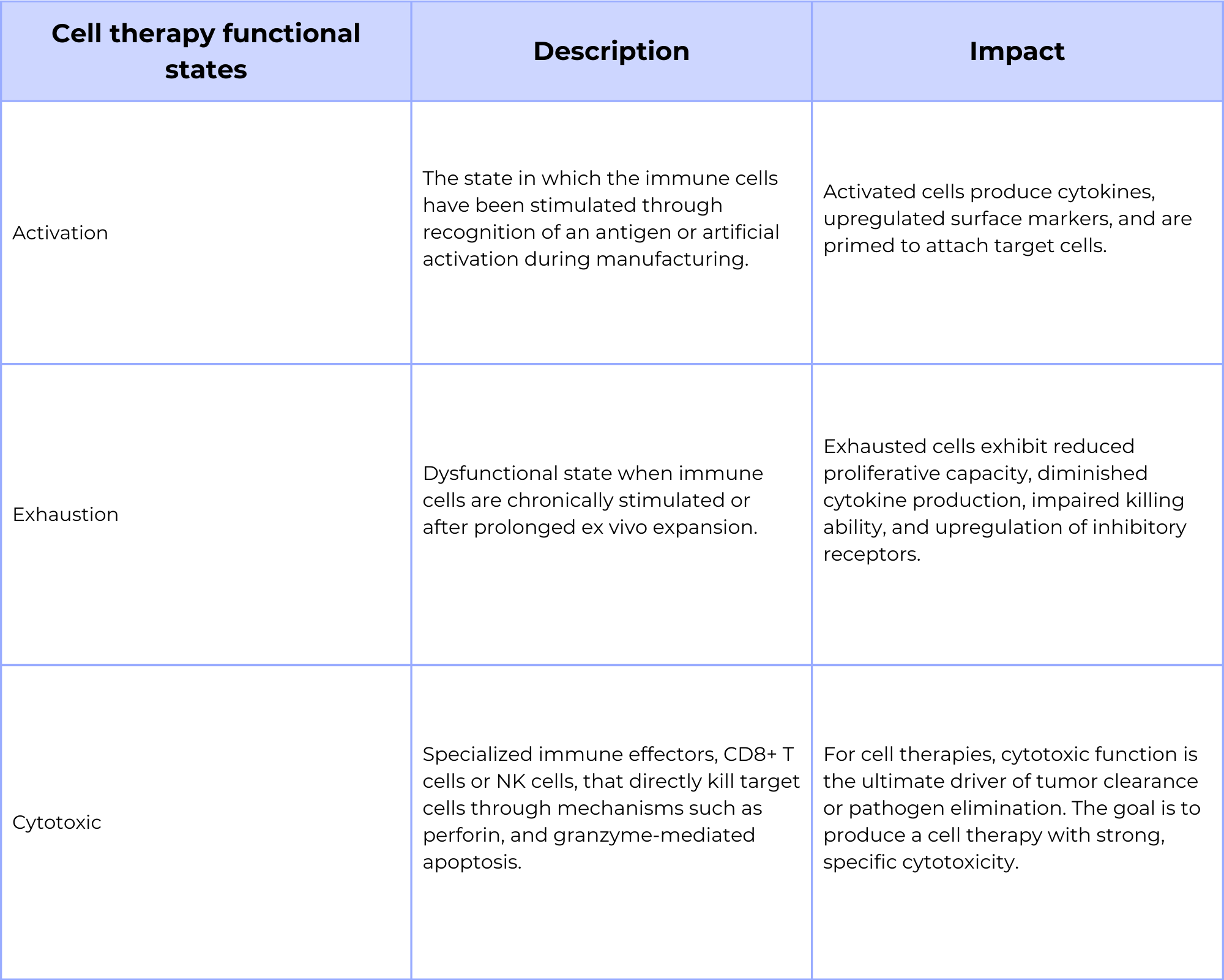
In therapies that incorporate immune cells in the therapy, like CAR-T cells, the functional properties of the final product are directly tied to the diversity of the T-cell receptor (TCR) or B-cell receptor (BCR) clonotypes. Single-cell multi-omics assays can simultaneously profile the clonotype and the gene expression of thousands of individual cells. This enables researchers to connect specific clonotypes to a cell’s transcriptional state, revealing variability in the final therapy product, including the surface protein and receptor function. For example, single-cell multi-omics can help identify if certain clonotypes are prone to exhaustion or activation, providing a crucial, per-cell understanding of therapeutic product’s efficacy that is lost in bulk analysis.
Identify cell therapy exhaustion, activation, and cytotoxicity through gene expression signatures
Beyond simple clonotype-function relationships, single-cell multi-omics methods that incorporate targeted gene expression profiling can identify subtle, yet critical transcriptional signatures within the cell therapy. When developing cell therapies like CAR-Ts, these gene expression signatures can indicate a cell’s functional state, such as whether it is activated, exhausted, or cytotoxic. By identifying these signatures, researchers can characterize the different cell subpopulations within the therapeutic product. This allows for the development of strategies to isolate or purify highly potent subpopulations, optimize manufacturing protocols, and ultimately enhance the efficacy and safety of the final cell therapy.
Table 2: Functional states of cell therapies
Correlating cell therapy vector copy number (VCN) with transgene expression
When developing cell therapies and rely on bulk analysis methods, those methods only provide an average of the vector copy number (VCN) across a population. This masks the cell-to-cell variability that can critically impact both safety and efficacy. Single-cell multi-omics analysis overcomes this limitation by simultaneously measuring VCN and gene expression in individual cells. This approach reveals how variation in vector integrations affect both transgene expression and endogenous gene regulation, enabling the identification of high risk clones with excessive integrations and the assessment of a cell’s functional state at the single-cell level for the cell therapy. Linking VCN to transcriptional gene expression, single-cell multi-omics provides a detailed, actionable view of the cell therapy’s performance that bulk assays cannot capture.
Table 3: Benefits targeted single-cell DNA and RNA profiling in cell & gene therapies
Mission Bio’s Pharma Assay Development (PAD) services is a collaborative partnership
Developing robust single-cell multi-omics assays for your drug and therapy program, particularly those that need to capture gene expression, gene fusion, and transcriptional states, require both specialized expertise and a tailored approach. Mission Bio’s Pharma Assay Development (PAD) services provide a custom, fit-for-purpose service designed to meet the specific needs of your program. Instead of providing a one-size-fits-all service, PAD serves as a strategic partnership, working closely with you to design studies and address key questions that generate the most relevant data across all stages of your program development, from early discovery and preclinical studies to IND submission and clinical trials.
This fit-for-purpose service begins with a comprehensive project scope and custom panel design. The Mission Bio team can assist in defining panel targets and collaborate on sample selection to generate data that is both technical robust and clinical relevant. Using the Tapestri Platform, our expert team performs single-cell multi-omics data collection and analysis, resulting in a comprehensive report. By partnering with Mission Bio, you can de-risk your programs, leveraging our deep experience in single-cell multi-omics and its advantages to generate the supporting data needed to make informed decisions throughout your drug or cell therapy development, increasing the likelihood of your success in the clinic. Mission Bio’s support does not stop at just developing your assays, transferring them to scale for commercialization is just as important.
Scaling single-cell multi-omics assays for clinical and commercialization
As a therapeutic program progresses from early development to clinical trials and eventual commercialization, the need for scalability, standardized processes, and regulatory compliance become paramount. Most biotech and pharmaceutical companies developing therapies rely on external partners like contract research organizations (CROs) and contract development and manufacturing organizations (CDMOs) to provide the necessary infrastructure and expertise to scale efficiently. CROs typically specialize in preclinical and clinical trial management, while CDMOs offer end-to-end support that combine development and manufacturing services to easily transition from early development to commercial production.
When you develop an assay that validates your therapy successfully and need it to be used in Good Manufacturing Practice (GMP) or clinical settings, partners who have experience in this space will make the transition easy and less frustrating. Mission Bio’s PAD services are designed to facilitate this exact transfer process. The PAD team works to ensure that the custom assay you collaboratively developed can be successfully and reproducibly run at the partnering CRO or CDMO. This includes providing the necessary standard operating procedures (SOPs), bioinformatic pipelines, and formal documentation to support the assays’ integration into the drug development and manufacturing workflow. By offering a clear path for assay transfer, Mission Bio can help scale successfully.
Fit-for-purpose single-cell multi-omics for therapy success
In oncology and cell therapy programs, targeted single-cell DNA and RNA profiling, combined with gene expression analysis, links gene mutations to transcriptional and functional states at the individual cell level. This enables the dissection of tumor or therapy product heterogeneity, the identification of rare subpopulations, and the direct assessment of gene edits. Mission Bio’s Pharma Assay Development (PAD) services make this achievable through customizable, fit-for-purpose services, from study design and panel selection to sample processing, gene expression and multi-omics analysis, and comprehensive reporting. PAD services support your program across all stages, from early discovery to clinical trials, including assay transfer for commercialization. By partnering with Mission Bio, you can de-risk your program and ensure single-cell multi-omics assays provide the insights needed to ensure your therapy programs success in the clinic.










