Genome engineering techniques, like CRISPR, have revolutionized how scientists approach research and precision therapeutics. While a powerful technique, especially for engineering novel therapeutics, CRISPR introduces some inherent challenges related to heterogeneous editing outcomes that include variations in zygosity, off-target effects, and co-occurring multiplex edits. Here we explore these variations, including how they occur and how single-cell multi-omics technology can help identify and quantitate these outcomes.
How do CRISPR-edited pools become heterogeneous?
Despite the promise of precision of advanced genome editors, editing outcomes remain largely unpredictable. Different cells subjected to the same editing regimen can yield distinct combinations of edits, varying not only across multiple on-target sites but also between on-target and off-target locations. The result is that genome editing of pools is often very heterogeneous.
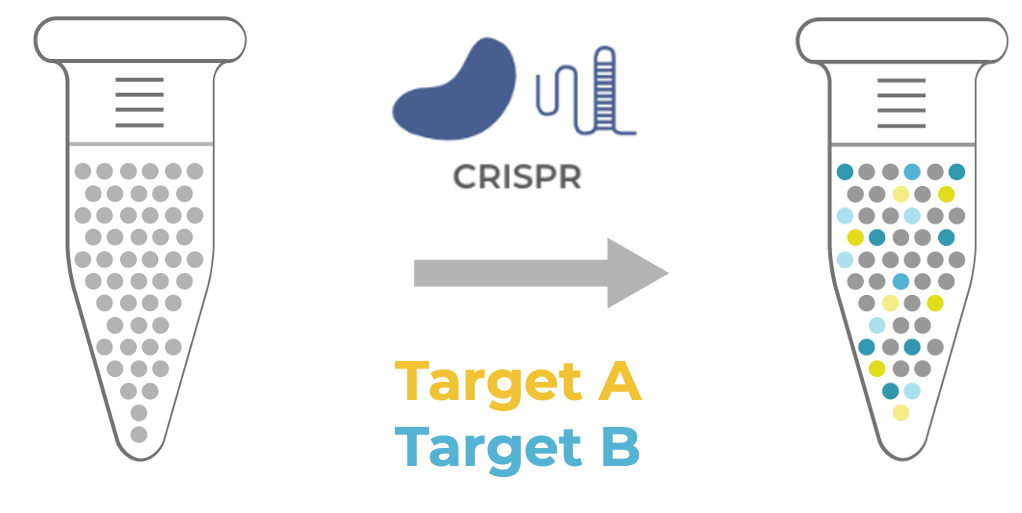
Figure 1. CRISPR editing of two intended targets (A and B) leads to heterogeneous outcomes, in which the edit genotypes, zygosity status, and co-occurrence vary across cells (illustrated by gradation in color)
Figure 1 shows an example of an editing experiment where there are wild type cells that are genome-edited using CRISPR at two intended targets, Target A in yellow and Target B in blue. After editing, the result is a mixture of cells, some of which are edited with the editing shown as a gradation of the blue and yellow colors to represent the variability in the pool.
What are the types of variation in CRISPR-edited cells?
Variation in zygosity
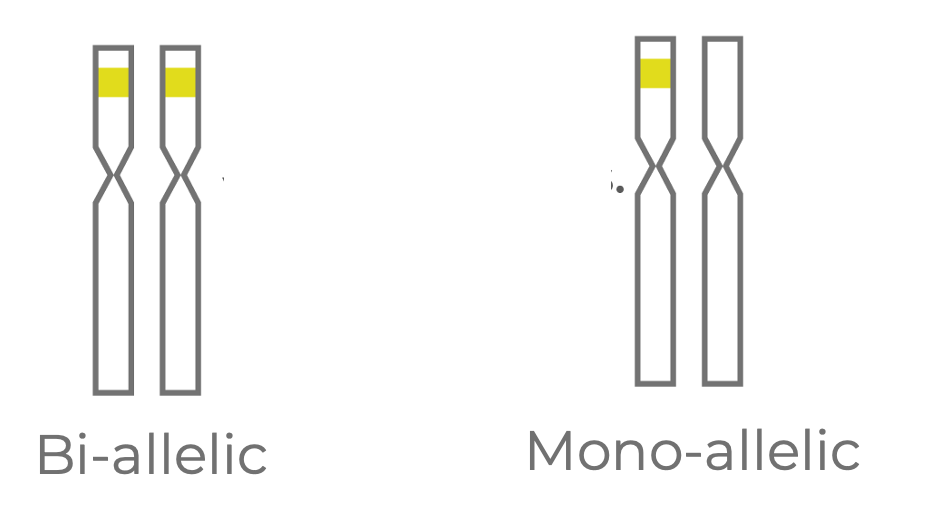
Figure 2. Edits at an intended target may be bi-allelic (present on both homologous chromosomes of a diploid cell) or mono-allelic (present on just one chromosome).
One source of variation is at the level of zygosity status (Fig 2). In our example, let’s assume the cells are diploid and that two homologous chromosomes can be edited for each target site. You could have a bi-allelic edit where both chromosomes are edited, or you could have a mono-allelic edit where one chromosome is edited and one chromosome remains wild type.
Variation in variant genotype
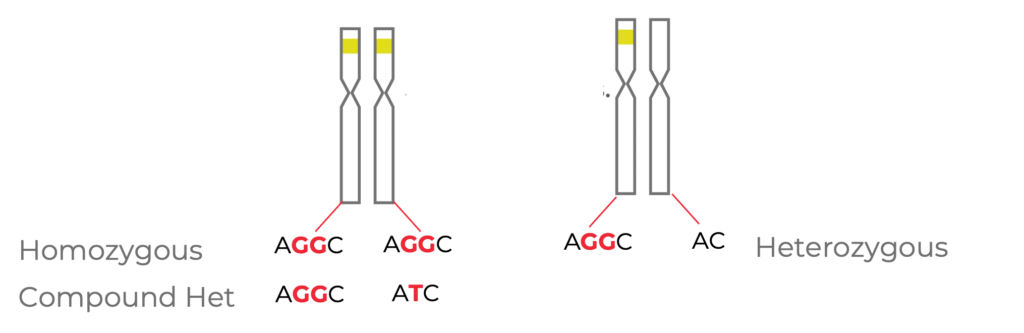
Figure 3. Cells that are edited with the same guide RNA at the same target site may have different variant genotypes. Cells may be homozygous, compound heterozygous, or heterozygous at the target locus.
If we dig into this further, more variation is possible. In the example above (Fig. 3), for a bi-allelic edit (left), one option is a homozygous edit, where the same edit occurs on both alleles (in this case, an insertion of two G nucleotides).
Another possibility is a compound heterozygote, where both chromosomes are still edited, but there are different indels on each of those chromosomes (in this example, the insertion of two G nucleotides in the first, and the insertion of a T nucleotide in the second).
Finally, you can have a heterozygous edit, where one chromosome is wild type and one is edited (insertion of two G nucleotides).
Variation in co-occurrence of edits
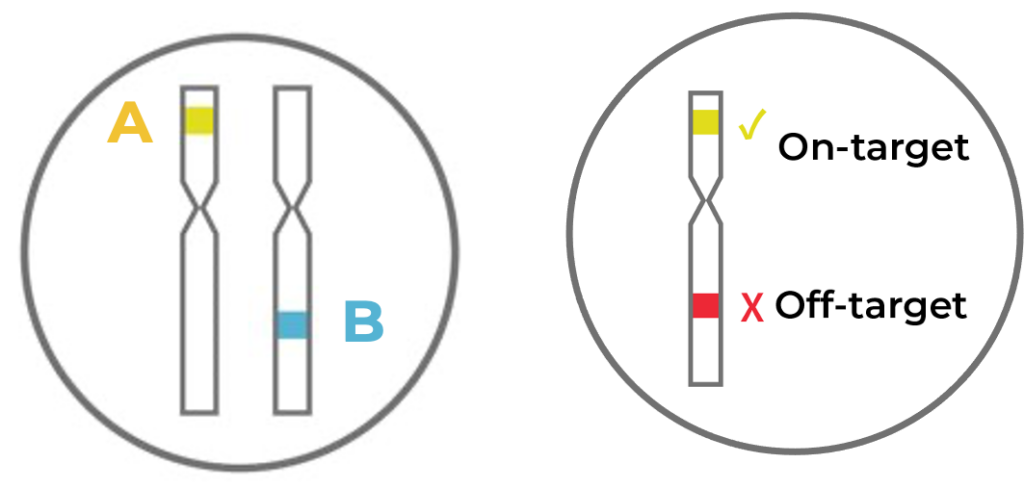
Figure 4. When editing multiple intended target loci at the same time (illustrated as A and B on the left), not all cells will have both A and B targets edited.
Because our experiment involves editing multiple targets (A and B), variation can also exist on how many targets are actually edited across cells. Some cells might have both targets edited at the single cell level, but some cells might just have one target edited (Fig 4, left) If off-target edits are present, there might be different combinations of on- and off-target edits that also vary across cells (Fig 4, right).
Variation in cell types or states that are edited
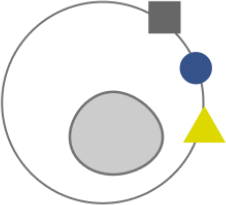
Figure 5. Cell types/states can be identified by the signature of cell-surface proteins. Gene editing may differentially edit some cell types/states more readily than others, leading to cell-to-cell variation in editing outcomes.
Finally, if we assume the cells in our experiment differ in cell type or cell state, editing may differ across these categories. So this is an additional axis of variation that must be considered.
It’s important to note that while bulk sequencing is appropriate for some questions, bulk assays do not preserve the information of individual cells and can’t answer these questions in variation outlined above. Only single-cell analysis can provide insight into these levels of variation.
The Tapestri Genome Editing Solution offers comprehensive analysis of gene edits
So how do we address all these questions in variation? The Tapestri Platform can be used for a diverse array of applications ranging from assessing tumor heterogeneity, all the way to cell characterization for genome editing and cell and gene therapies, as well as disease modeling.
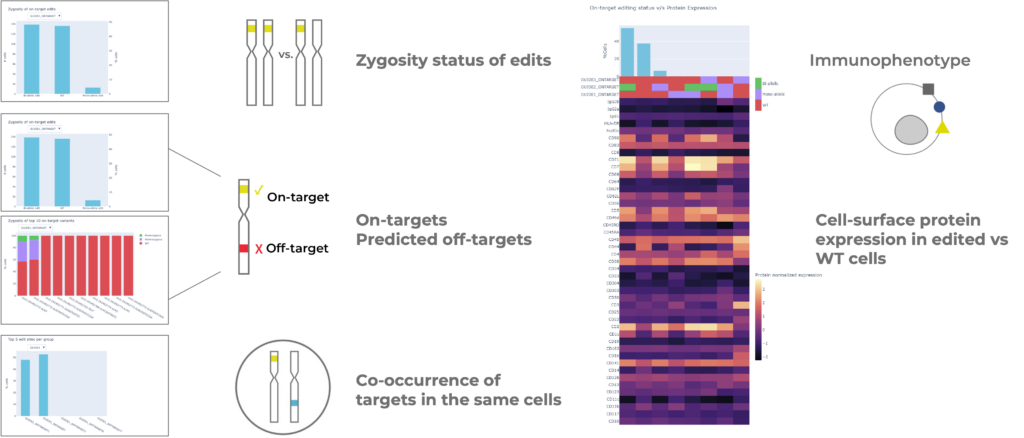
The Tapestri Genome Editing Solution is an end-to-end solution that includes reagents, the Tapestri instrument, and a robust software solution for analysis. This assay is high throughput and can analyze 1000s of cells at a time, unlike plate-based assays which are restricted to fewer cells. The assay is also targeted, meaning the whole genome does not need to be sequenced.
The Tapestri Genome Editing Solution addresses:
- On-target single & multiplexed editing. If you are editing one or multiple sites simultaneously, the solution can analyze all these targets together.
- Predicted off-target editing. If you know the predicted genomic location of the off-target edits, the solution can confirm if off-target editing is occurring. There are multiple tools to predict off-target edits prior to a Tapestri assay.
- Edit zygosity. The analysis will tell you whether one or both chromosomes are edited at each target site and the identity of the variants.
- Edit co-occurrence. The analysis will report the co-occurrence of edits at multiple intended targets (on-targets) and the co-occurrence of on- and off-targets in individual cells.
- Multi-omic analysis. Cell surface proteins can be co-measured with genome edits to provide insights into the immunophenotype of edited cells or to validate the knockout of a target.
To learn more about how to analyze heterogeneous outcomes in genome-edited pools, check out our joint webinar with Synthego or download our Genome Editing Solution Application Note for more information.