Cancer is a Tough Nut to Crack
Despite decades of research and therapeutic development, cancer still remains the second leading cause of mortality in the USA. The difficulty with this disease lies in its complexity. Tumors are composed of subpopulations of cells — or clones — that each has a unique collection of mutations. It is this heterogeneity coupled with the capacity to evolve that gives tumors their ability to evade therapies. For instance, if a given clone has a mutation that confers resistance to a drug, then it will survive even though neighboring cancer cells die. Even if the tumor is seemingly eliminated, the resistant clone will likely continue to multiply and cause relapse at a future time point.
Single-cell Multi-omics Enables a High Resolution Look into Cancer
Despite the challenges with treating cancer, the rapid development of genomics technologies over the past few years has brought new hope. One such advancement has been single-cell multi-omics, in which multiple analytes are measured in individual cells. Mission Bio’s Tapestri Platform enables robust DNA + protein multi-omics at the single-cell level. In a streamlined workflow, researchers can measure single-nucleotide variants (SNVs), copy number variants (CNVs), and cell-surface proteins in the same cells.
Why is it important to measure both DNA and protein at the single-cell level? The answer is that tumors are both genotypically and phenotypically heterogeneous. They comprise cells that harbor different combinations of mutations and also differ by disease state, lineage, and stage of maturity.
Measuring DNA variation at the single-cell level enables researchers to dissect the clonal architecture of tumors (something bulk NGS cannot do with certainty). Measuring the collection of cell-surface markers — or immunophenotype — of cells imparts information about cell lineage and state. Measuring both the genotype and immunophenotype of each cell thus gives researchers a more vivid picture of a tumor.
Insights from multimodal studies not only add to the foundational knowledge of cancer but also contribute to therapeutic development and clinical strategies. Here are some ways that multi-omics information is used:
- Biomarker identification: identifying new biomarkers of cancer can be used for diagnosis, prognosis, and patient stratification.
- Minimal residual disease (MRD): identifying whether a small number of cancer cells are still present in patients following treatment. Detecting these cells is important because they may cause relapse.
- Drug targeting: cell-surface proteins can act as targets for drugs so that cancer cells can be effectively targeted and destroyed [1,2]. Measuring both genotype and immunophenotype ensures that all malignant cells (determined by a diagnostic aberrant genotype) also carry the marker that is targeted.
- Identifying CHIP: for patients with leukemia, distinguishing malignant cells from benign ones can be challenging. As humans age, hematopoietic stem cells tend to accrue mutations that make them clonally expand, a phenomenon called clonal hematopoiesis of indeterminate potential (CHIP). CHIP clones are not malignant but have some of the same mutations as leukemic clones, making them difficult to distinguish. Combined single-cell genotypic and immunophenotypic information can better enable clinicians to determine whether patients indeed have leukemia [3].
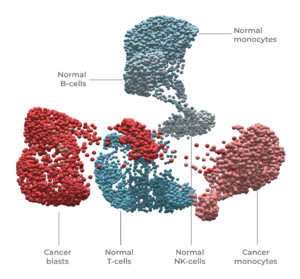
Single-cell DNA + protein multi-omics was used to resolve and state (normal vs cancer) and cell type (T-cell, B-cell, blast, monocyte, NK) in this leukemia sample.
Interested in how single-cell multi-omics is used in cancer research? Sign up to hear Dr. Ross Levine on Wed, April 21, 1pm ET (10am PT).
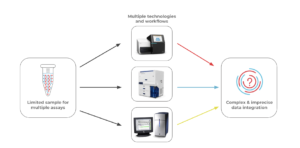
Conventional Multi-assay Workflows Present Challenges
The multi-omics approach is powerful in that it provides a detailed look at multiple types of cellular information. Traditional workflows that measure genotype and immunophenotype often involve bulk NGS (genotyping), flow cytometry (immunophenotyping), and cytogenetic analysis (for CNV) as separate assays. Using a multi-assay approach, however, doesn’t come without drawbacks. Here are a few:
- Clinical samples are often small in size and difficult to obtain, so splitting a sample across multiple assays leaves little for downstream analyses.
- Data integration from multiple assays is often a complex process that requires bioinformatics expertise.
- Multiple assays on different instruments can be expensive and often involves long timelines before researchers get answers to their questions.
- Conventional workflows either utilize bulk measurements or cannot be easily set up to measure individual cells in a high-throughput manner.
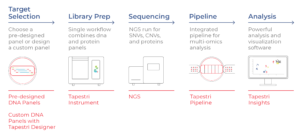
Tapestri Delivers Multimodal Data in a Single Assay
Mission Bio’s Tapestri Platform analyzes the DNA and cell-surface proteins in 1000s of individual cells at a time. In a streamlined workflow, users first select a DNA panel with their desired targets. The sample is stained with a protein panel (comprising antibody-oligo conjugates, or AOCs, that bind specifically to target cell-surface proteins) and deposited into the Tapestri instrument. Individual cells are then encapsulated in droplets, where target genes and AOCs are amplified with a cell-specific barcode and prepped for NGS. After sequencing, the data are processed and analyzed using Tapestri software.
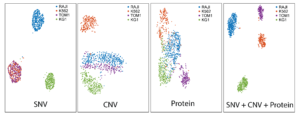
Single-cell DNA+ protein analyses yield detailed datasets that enable robust cellular classifications. The dataset below (t-SNE projections) shows groupings of four cell lines based on SNV, CNV, and cell-surface protein information. The plot that utilizes all three parameters (far right) portrays the most distinct clusters, illustrating how multi-modal analyses can reveal classifications that are not apparent with a single analyte.
Making Headway Against Cancer, One Cell at a Time
The use of DNA + protein multi-omics has recently led to significant insights into the evolution of heme malignancies, like acute myeloid leukemia (AML). In an impactful study recently published in Nature, Ross Levine (Memorial Sloan Kettering Cancer Center) and his colleagues used single-cell multi-omics to elucidate the process of leukemic transformation [4]. The team found that clonal hematopoiesis CH (a pre-leukemic state that involves initial mutations seen in AML) and full-blown AML had distinct genotype-immunophenotype associations. They also found that the addition of mutations within clones, especially in signaling genes, changed clonal immunophenotypes.
Want to hear about this Nature study? Sign up for Dr. Levine’s webinar on April 21, 1pm ET (10am PT).
Several other studies have capitalized on Tapestri-enabled multi-omics to study cancer. For instance, Morita et al. (2020) reported phenotypically aberrant populations in relation to the stepwise acquisition of AML driver mutations [5]. These insights contribute to our understanding of pathogenesis in the context of hematopoiesis. Catherine Smith (University of California, San Francisco ) and her colleagues investigated the genotype-immunophenotype relationships of AML clones in different clinical scenarios [6]. Using single-cell multi-omics, the team demonstrated that while the association between genotype and immunophenotype was concordant in some patients, it was decoupled in others. This study illustrates the complex genotype-immunophenotype relationships in AML and underscores the value of measuring these parameters together in order to accurately characterize tumors.
See Catherine Smith discuss this study. Sign up for the webinar on May 6, 1pm ET (10am PT).
Marching Towards a Future of Precision Medicine
Advances in single-cell multi-omics hold promise in the translational space, where basic cancer research can inform clinical practice. With the ability to scrutinize cancer with greater resolution, clinicians are better positioned to make accurate diagnoses and prognoses and even tailor therapies based on unique tumor profiles. Even though cancer is a tough nut to crack, tools like single-cell multi-omics are proving to be the “nutcrackers” scientists need to make progress. Combining greater resolution with enhanced personalization will likely bring about great change in how cancer is studied and treated — for the better.
Has single-cell analysis caught your fancy? Check out our introductory ebook!
References
- Buckley S A & Walter RB. Antigen-specific immunotherapies for acute myeloid leukemia. Hematology 2015, 584-595 (2015).
- Andrews TE et al. Cell surface markers of cancer stem cells: diagnostic macromolecules and targets for drug delivery. Drug Deliv Transl Res. 2013 Apr;3(2):121-42.
- Dillon LW et al. Personalized single-cell proteogenomics to distinguish acute myeloid leukemia from non-malignant clonal hematopoiesis. medRxiv 2020.10.22.20216028.
- Miles LA et al. Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature. 2020 Nov;587(7834):477-482.
- Morita K et al. Clonal evolution of acute myeloid leukemia revealed by high-throughput single-cell genomics. Nat Commun. 2020 Oct 21;11(1):5327.
- Demaree B et al. Joint profiling of DNA and proteins in single cells to dissect genotype-phenotype associations in leukemia. Nat Commun. 2021 Mar 11;12(1):1583.











