What is CHIP?
Hematopoietic stem cells (HSCs) are multipotent cells that give rise to differentiated blood cells including red blood cells, white blood cells, and platelets. HSCs are found in umbilical cord blood, peripheral blood, and adult bone marrow. These cells are able to self-renew (i.e., replicate while remaining in an undifferentiated state). However, such divisions are rare, enabling the genome to remain quite stable.
Despite the genetic stability of HSCs, these cells gradually acquire somatic mutations as humans age. Frequently mutated genes include the epigenetic regulators DNMT3A, TET2, and ASXL1, collectively referred to as DTA mutations (incorporating the first letter of each gene). HSCs with these mutations have a competitive advantage, such that they are able to self-renew more often than HSCs without the mutations. This leads to a condition called clonal hematopoiesis of indeterminate potential (CHIP), in which hematopoietic stem cells (HSCs) clonally expand into subpopulations.
CHIP is an age-related condition, with DTA mutations begin accumulating in HSCs around middle age (Fig 1). Approximately 20% of the US population have this condition, but most are not aware of it. CHIP is usually diagnosed when a blood or bone marrow test reveals an associated genetic mutation DTA mutations occurring at a variant allele frequency (VAF) of at least 2% without cytopenia (reduction of blood cells).
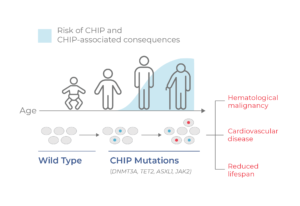
Figure 1. CHIP progresses with age. CHIP is a non-malignant condition characterized by mutation and clonal expansion of blood cells, beginning at middle age. CHIP is associated with a higher risk of developing hematological malignancies and cardiovascular disease, as well as a reduced lifespan.
A Pre-Malignant State, but Not Cancer
CHIP is nonmalignant and does not present any symptoms. However, people with CHIP have a higher risk of developing blood cancer, such as leukemia. This is because the DTA mutations that characterize CHIP are also early-stage mutations typical of heme malignancies like myelodysplastic syndromes (MDS) or acute myeloid leukemia (AML). The “indeterminate potential” part of the name CHIP refers to the uncertainty of the fate of clonal populations of stem cells— whether they will transform into cancer or not.
In addition to increased risk for heme malignancies, people with CHIP mutations are also at increased risk of developing cardiovascular disease and inflammation-related reduced lifespan (Fig. 1). There is also evidence that people with CHIP who receive treatment for other types of cancer may be at higher risk of developing leukemia.
Prognostic Value of CHIP
One reason why CHIP is important is its potential prognostic value. Active areas of research include whether CHIP can be used to inform when clonal hematopoiesis becomes a medical event, including the risk of developing hematological malignancy. Because DTA mutations are found in both nonmalignant (CHIP) and leukemic cells, CHIP blurs the line between healthy and diseased states. Though investigation in this area is still in its infancy, there is potential to use CHIP to identify at-risk individuals so that therapeutic intervention may be taken before cancer (or cardiovascular disease) develops.
CHIP and Minimal Residual Disease
Another area of significance for CHIP is the identification of minimal/ measurable residual disease (MRD) — the small quantities of cancer cells that remain following treatment. After AML patients undergo treatment and reach complete remission, the presence of MRD has strong predictive power of relapse post-therapy and is integral to forming a plan for post-treatment care.
Yet, the presence of CHIP complicates the detection of MRD. Because DTA mutations are present in both CHIP and AML clones, it may be difficult to distinguish the two. In addition, leukemic clones may evolve from pre-existing CHIP clones, or they may arise independently. Understanding the clonal architecture and evolution through periods of treatment periods will help researchers (and potentially clinicians) to identify true MRD from age-related CHIP. It remains an unmet need to resolve the subclonal biology to improve clinical management. That is where single-cell DNA sequencing (scDNA-seq) and multi-omics come in.
Distinguishing CHIP and MRD with Single-cell Analysis
Conventional methods of MRD detection include bulk next-generation sequencing (NGS) and flow cytometry. Bulk NGS measures the presence of DTA mutations in a cell population. However, since this method combines DNA across cells, it cannot distinguish whether DTA mutations reside in CHIP clones, in leukemic clones, or both. Another method of MRD identification is flow cytometry, which assesses cell-surface markers to identify aberrant immunophenotypes. However, this method is often discordant with bulk NGS and may not catch all genetically mutant cells. Moreover, differences in immunophenotype of CHIP vs leukemic clones have not been extensively defined.
The solution to enhanced MRD detection may involve the assessment of samples at the single-cell level. Single-cell DNA sequencing (scDNA-seq) on Mission Bio’s Tapestri Platform can be used to resolve the clonal architecture of samples, and by doing so, distinguish CHIP from malignant clones (Fig. 2). For instance, Ediriwickrema et al. (2020) used scDNA-seq to evaluate AML samples at diagnosis, remission, and relapse. Through their analysis, they were able to quantify co-occurring variants, differentiate clonal hematopoiesis from relapse-causing clones, and identify clinically relevant MRD.
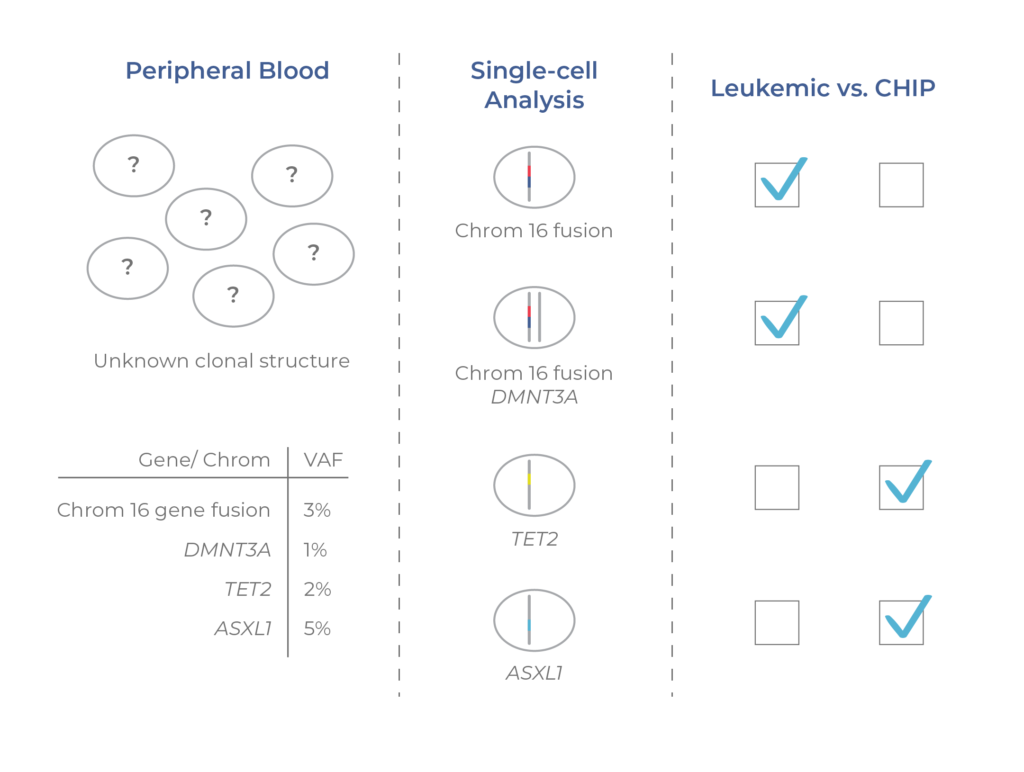
Figure 2. Single-cell analysis distinguishes CHIP clones. Pre-malignant (CHIP) and malignant clones cannot be distinguished from VAFs alone. By resolving the mutational structure across clones, scDNA-seq enables the identification of CHIP and leukemic clones.
In another study, Dillon et al. (2021) used Tapestri’s multi-omic capability (DNA and cell-surface proteins) to investigate the clonal architecture of relapsed AML patients. By looking at the genotype and immunophenotype of individual cells, the researchers were able to confidently distinguish CHIP and MRD. Additionally, the researchers were able to identify cases where AML arose independently and where AML developed from precursor CHIP cells. As these studies illustrate, a single-cell approach can be used to resolve the clonal complexity of patient samples and robustly distinguish cancerous cells from CHIP.
See a summary of the Dillon et al. paper here.
Conclusion
Given the clinical significance of CHIP, there is a growing interest in understanding how this condition arises and transforms into cancer. CHIP be useful to inform a person’s risk for developing leukemia and thus aid in developing personalized treatments that preempt the disease. Understanding CHIP is also relevant for patients who have undergone cancer treatment when it is important to determine whether MRD is present or not. The use of single-cell DNA sequencing and multi-omics can be used to reveal the clonal architecture of patient samples, providing visibility into the line between benign and disease states. With a better understanding of CHIP, there is great promise for better treatment strategies and improved clinical management.
References
Dillon L.W., et al. Personalized single-cell proteogenomics to distinguish acute myeloid leukemia from nonmalignant clonal hematopoiesis. Blood Cancer Discov. 2021 May 2:1-7. doi: 10.1158/2643-3230.BCD-21-0046.
Ediriwickrema A. and Aleshin A., et al. Single-cell mutational profiling enhances the clinical evaluation of AML MRD. Blood Adv. 2020 Mar 10;4(5):943-952. doi: 10.1182/bloodadvances.2019001181.











