

This panel was designed by Omar Abdel-Wahab Lab at Memorial Sloan Kettering Cancer Center and covers many of the recurrent gene mutations in chronic lymphocytic leukemia (CLL). The panel was featured in a study published in the New England Journal of Medicine, “Mechanisms of Resistance to Noncovalent Bruton’s Tyrosine Kinase Inhibitors.” The panel was used to uncover the clonal architecture behind resistance to pirtobrutinib, a new class of non-covalent BTK inhibitors. The study allowed the investigators to identify the co-occurring mutations within each subclone, providing a picture of how new subclones evolved from pre-existing ones in response to therapeutic selective pressure. In addition, the authors found that the resistance mutations to this new class of non-covalent BTK inhibitors could also confer resistance to other non-covalent and covalent BTK inhibitors.
REQUEST QUOTEAdvance your understanding of the genetic heterogeneity underpinning chronic lymphocytic leukemia (CLL) by targeting 35 genes with 273 amplicons for single-cell sequencing. The CLL Published Panel is designed to target the most commonly mutated hotspots in CLL.
DOWNLOAD DATASHEET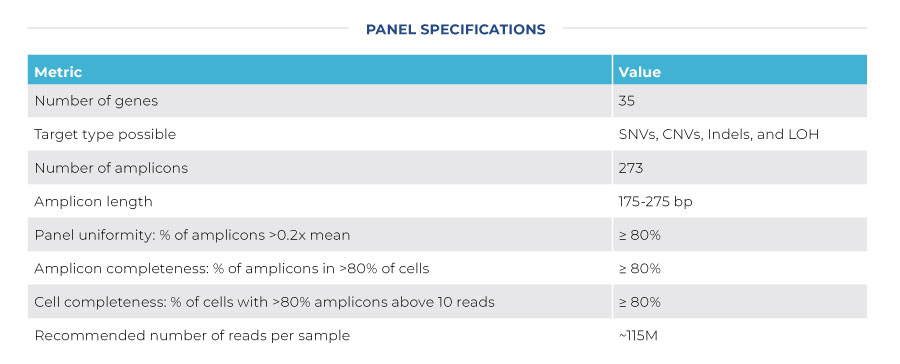

Use Tapestri Designer to add or remove content from existing catalog panels and make this panel your own with content that is relevant to your research. Or start from scratch and upload your human (hg19) or mouse (mm10) targets using gene names, genomic coordinates, or SNV IDs. Target anywhere in the whole human or mouse genome and advance your research with targeted single-cell DNA sequencing.
DESIGN PANEL