

The heterogeneity and dynamism of cancer present formidable challenges to understanding and treating the disease. As discussed in the last section, tumor evolution often leads to considerable genetic variation across clones. But the complexity of tumors does not stop at the level of DNA. Intratumoral phenotypic variation adds another layer of information that can be useful for research and clinical purposes. In this section, we discuss how single-cell DNA sequencing resolves variation at the genetic level, and how a powerful single-cell multi-omics approach correlates genotype and immunophenotype.
Deciphering the patterns of somatic mutations across clonal populations provides foundational insights into cancer. Traditionally, oncologists have relied on bulk next-generation sequencing (bulk NGS) to identify gene mutations present in tumor cells. This technique involves pooling the DNA across cells and measuring the variant allele frequencies (VAFs), i.e., the percentages of variant DNA molecules. Bulk sequencing is useful to confirm whether candidate genes are mutated and whether these mutations are germline (inherited) or somatic in origin. But because the cell identities are not preserved, bulk NGS is unable to measure patterns of co-mutation or resolve cell-to-cell variation in zygosity and copy number (Fig 4). This leaves researchers and clinicians with a black box when it comes to understanding the clonal diversity in their sample.
Fortunately, innovative single-cell DNA sequencing technology has now made it possible to elucidate the genetic heterogeneity of tumors. Sequencing may cover the entire genome or a subset of clinically relevant genes (gene panels). Although the former approach is appropriate for some objectives, targeted sequencing offers particular advantages for profiling tumor diversity. Because cancer continuously evolves, tumors often contain mutations that are relatively rare when they first emerge. Yet, detecting these rare mutations and the clones that carry them may be of significant clinical importance (e.g., minimal residual disease (MRD) and therapeutic resistance). In order to detect rare clones with confidence, high sequencing depth is necessary. Targeted DNA sequencing thus offers a cost-effective solution.
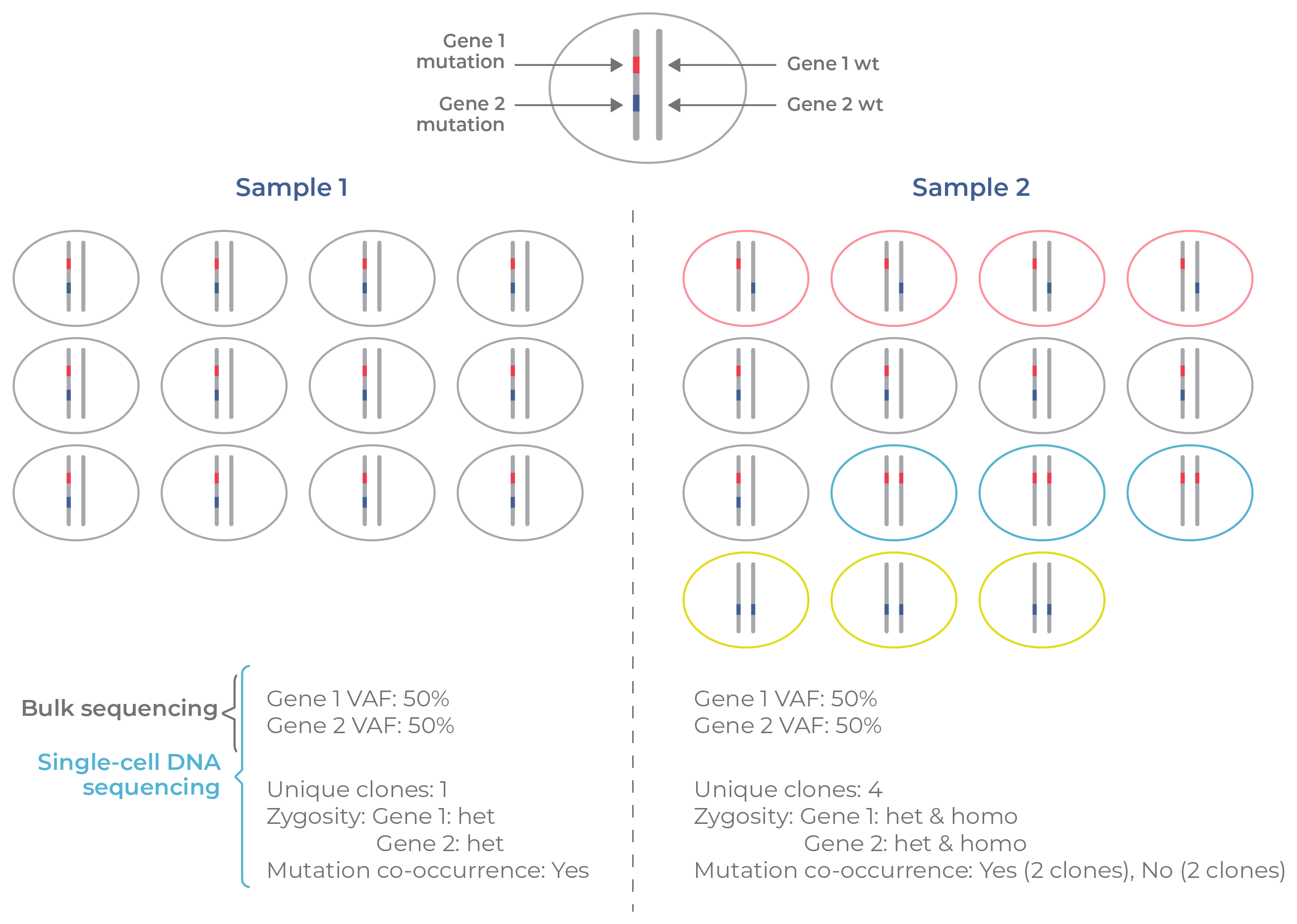
Figure 4. Bulk vs single-cell information on clonal structure. Consider two tumor samples (1 and 2) with clones that contain mutations in gene 1 (dark blue) and/or gene 2 (red). Sample 1 contains a single clone whereas sample 2 contains four unique clones (red, gray, blue, yellow) that differ in gene mutations and zygosity. According to bulk-calculated VAFs, both samples appear identical. Only single-cell sequencing identifies the differences in structure between the samples with regard to clone number, mutational co-occurrence, and zygosity.
Mission Bio’s Tapestri Platform enables highly sensitive targeted DNA sequencing at the single-cell level. By incorporating a cell-specific barcode to the amplified target sequences, this droplet-based technology measures genetic lesions (SNVs, indels, chromosomal rearrangements) and CNVs in each cell. Other structural characteristics like zygosity, mutational co-occurrence,and the presence of rare cell populations are also identified (Figs 4-5). This powerful technology has unlocked a host of new opportunities to understand how the mutational landscape of tumors fluctuates over time. But Tapestri’s capabilities do not stop there. Let’s next look at this platform’s powerful single-cell multi-omics capability.
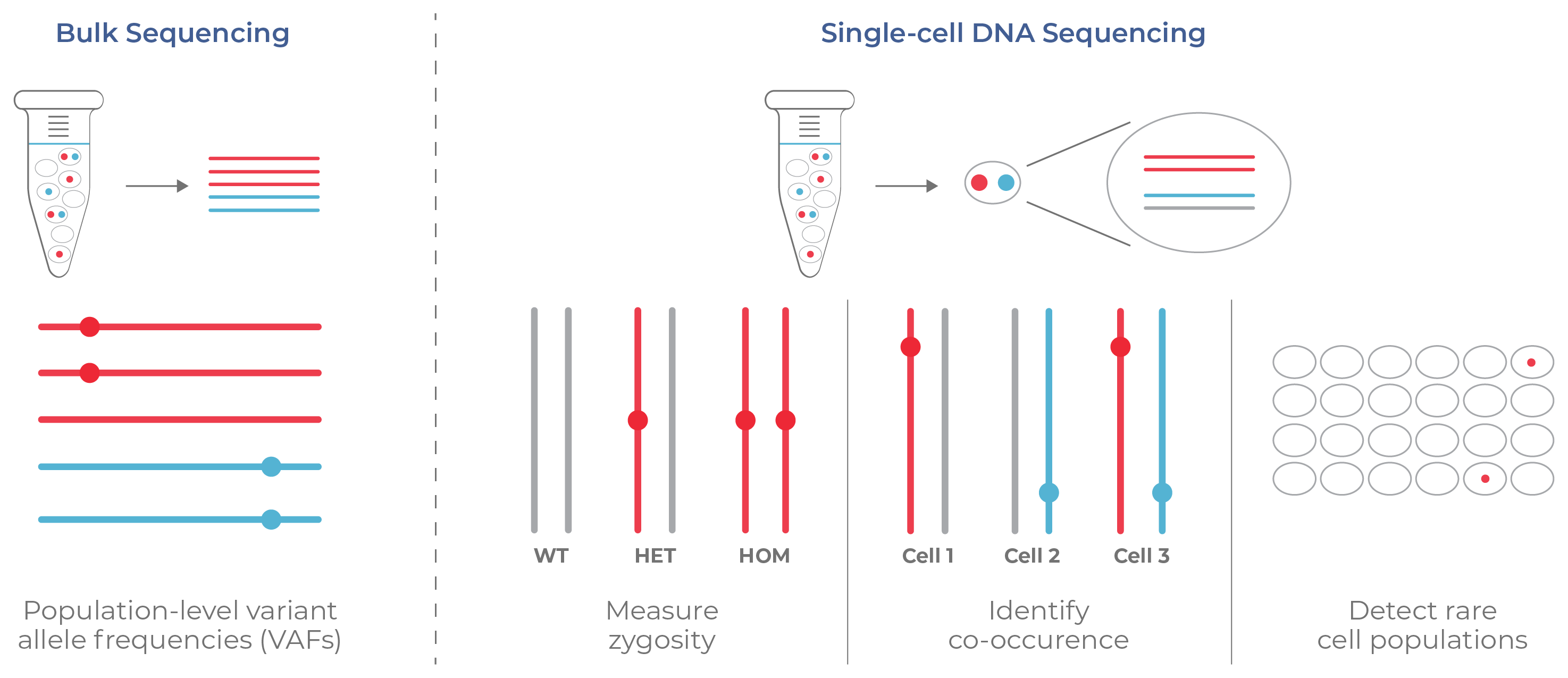
Figure 5. Information from bulk sequencing vs. single-cell DNA sequencing. Bulk NGS only provides an average readout of mutations (reported as variant allele frequencies, VAFs) and does not resolve the clonal structure of tumors. Alternatively, single-cell DNA sequencing assesses each cell and reveals the co-mutations and zygosity across clones. Rare cell populations are also detected.
Although measuring one parameter — like DNA — in a cell-specific manner provides an expanded view of cancer, a single analyte may not be sufficient to resolve all the clonal populations of a tumor. Single-cell multi-omics workflows, which simultaneously measure multiple analytes, often paint a more vivid picture. Combinations of different molecules, whether DNA, RNA, or proteins, facilitate the dissection of tumors at a more granular level.
Cell-surface markers are a particularly informative analyte. Adorning the plasma membrane of each cell, these antigens play important roles in cell signaling and immune surveillance. Their expression also correlates with certain cell characteristics like cell type, stage of development, and disease state. The marker profile for a given cell is called its immunophenotype.
When it comes to cancer, measuring immunophenotypes has considerable value. Tumors are not only genotypically heterogeneous but also phenotypically diverse. A given sample may contain healthy and pathogenic cells of different lineages and states. The simultaneous measurement of genotype and immunophenotype (i.e., proteogenomic profiling) classifies these cells into groupings that can be filtered and more closely investigated (Figs 6-7). Moreover, obtaining this information across thousands of cells portrays a thorough depiction of the tumor’s clonal diversity.
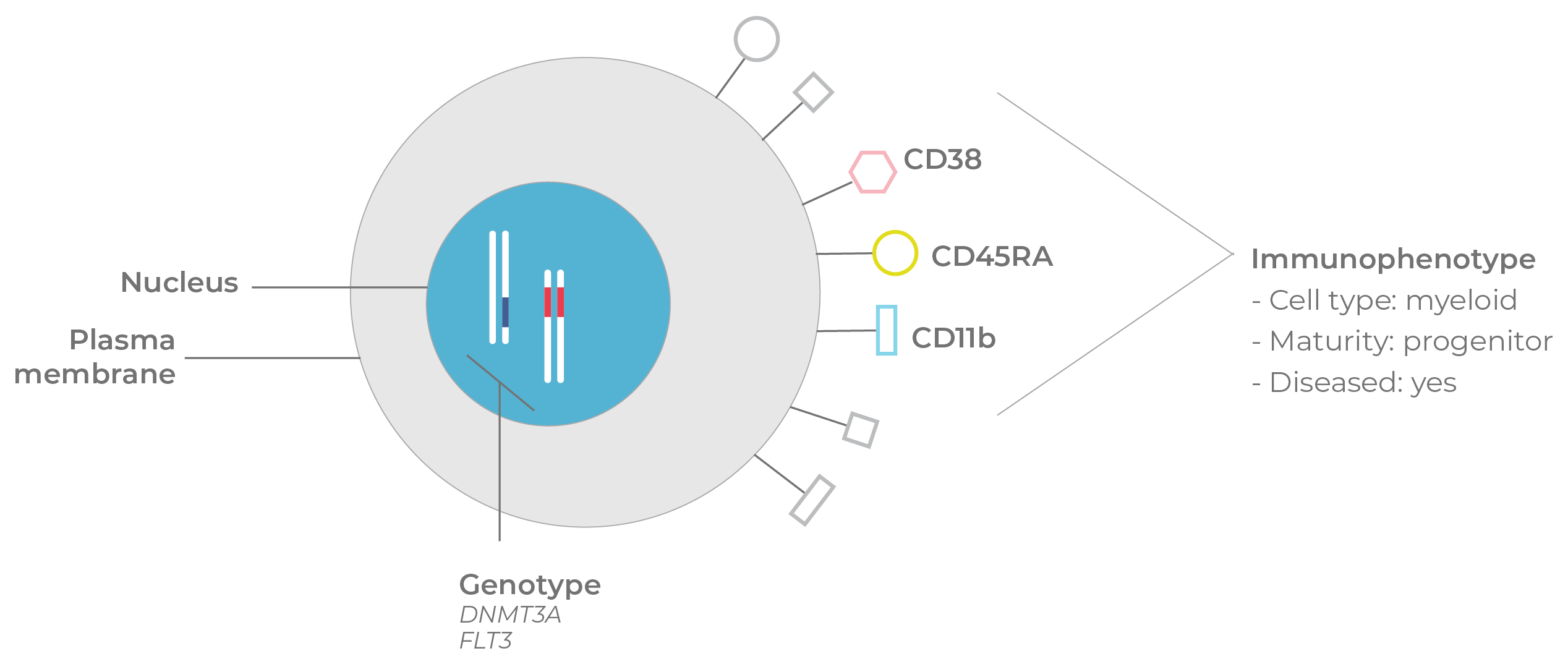
Figure 6. Genotype & immunophenotype. In this simplified example, information about mutations in cancer-associated genes (DNMT3A and FLT3) and immunophenotype suggests that the cell is a myeloid progenitor in a malignant state.
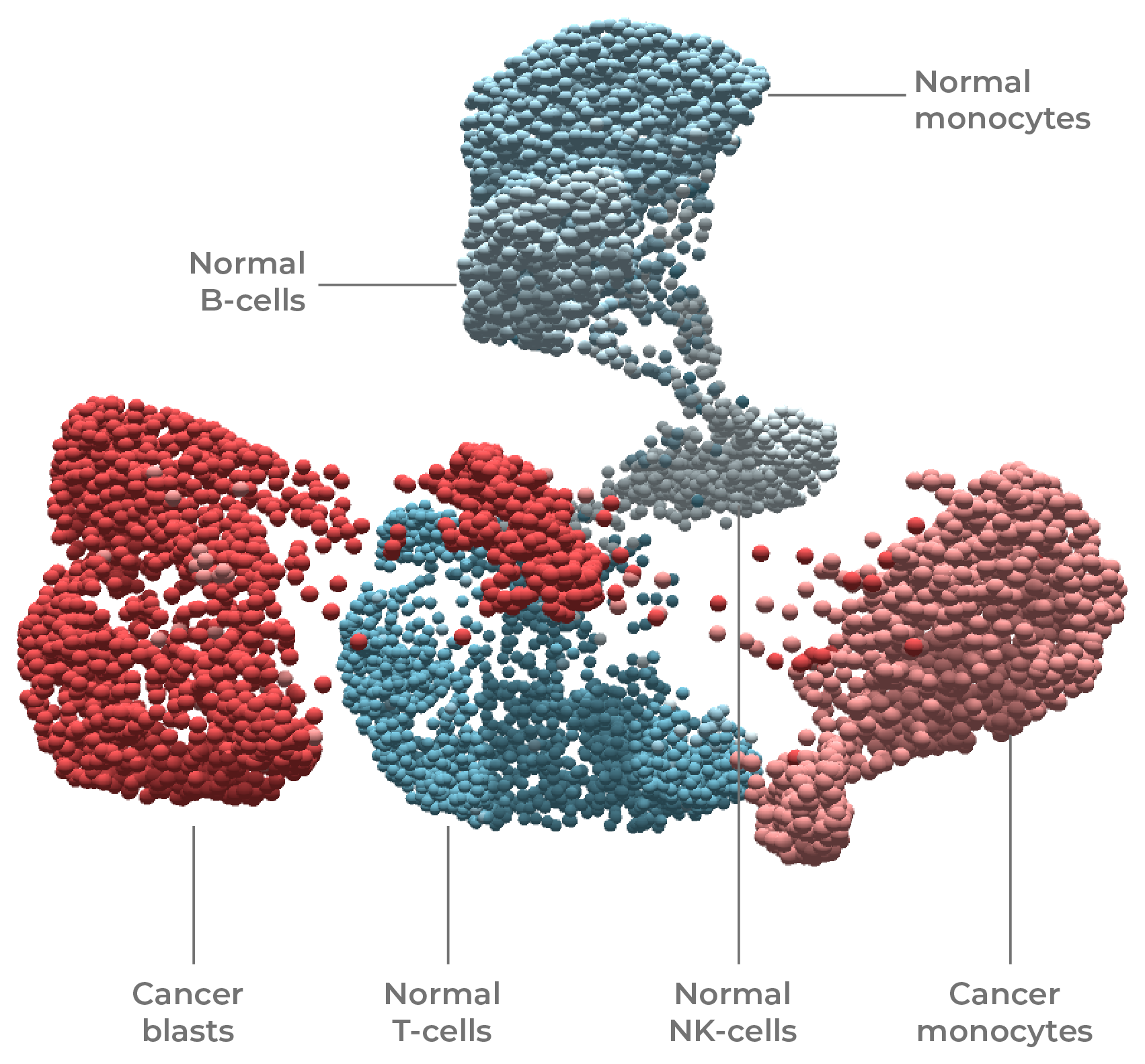
Figure 7. Tumors are genotypically & phenotypically complex. The use of DNA + protein multi-omics was used to resolve cell type (T-cell, B-cell, blast, monocyte, NK) and state (normal vs cancer) in this leukemia sample.
Conventional methods of proteogenomic profiling are laborious and require that samples be used in several assays across different instruments. In contrast, the Tapestri Platform enables high-throughput single-cell multi-omics (DNA + protein) in a single assay.
The single-cell multi-omics workflow begins with the selection of gene and protein panels. The former includes primers for targeted loci and the latter comprises antibody-oligo conjugates (AOCs) that bind specifically to target cell-surface proteins (Fig 9a). A short oligo attached to each antibody designates the identity of the corresponding antigen. The cell sample is first stained with a protein panel (preselected AOCs) and then deposited into a Tapestri cartridge where individual cells are encapsulated in oil droplets. Within each droplet, target genes and antibody-specific oligos are amplified with a cell-specific barcode. Libraries are then sequenced by NGS, and cell-specific mutational and protein profiles are reconstructed using Tapestri software (Figs. 8, 9b).
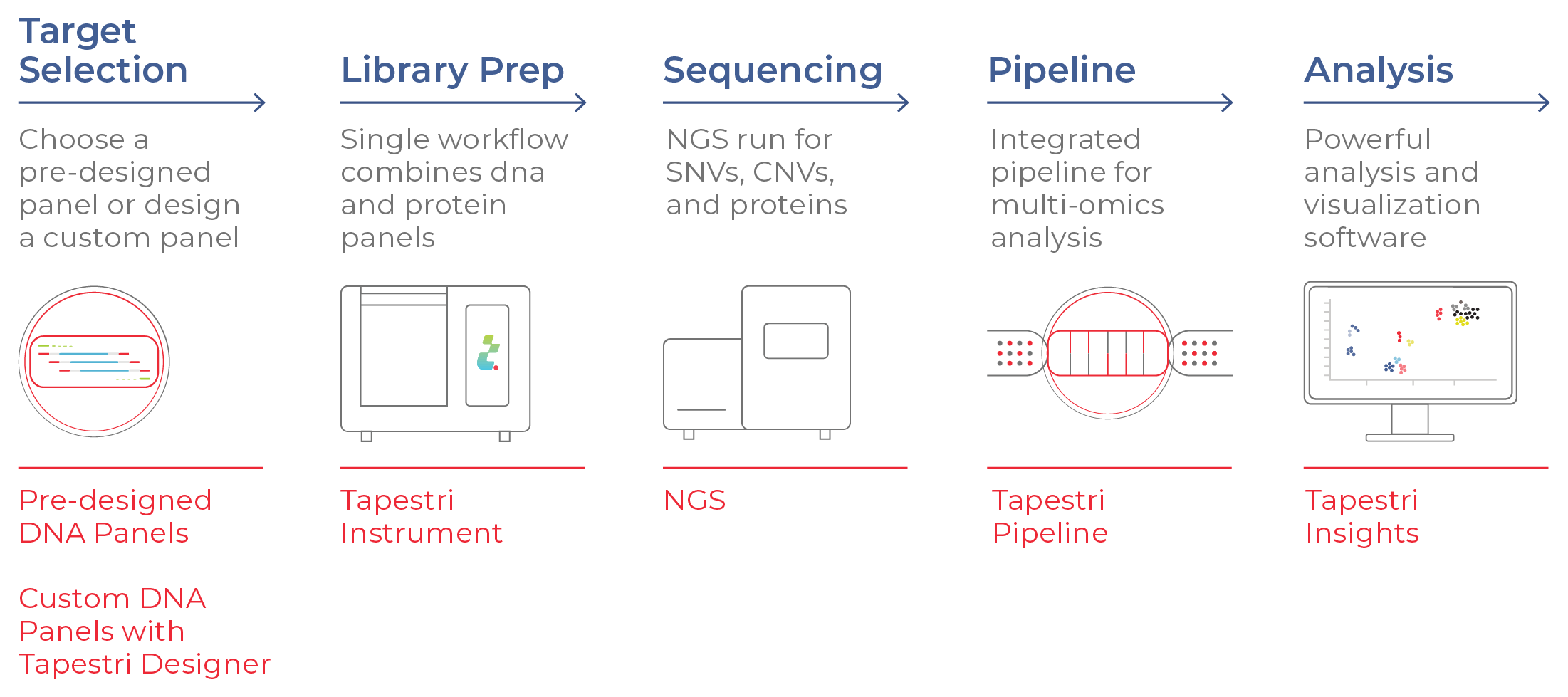
Figure 8. Single-cell DNA + protein multi-omics workflow. The workflow begins with the selection of DNA and protein panels, followed by target amplification and library preparation on the Tapestri Platform. NGS yields DNA and protein data that are then reconstructed and visualized at single-cell resolution.
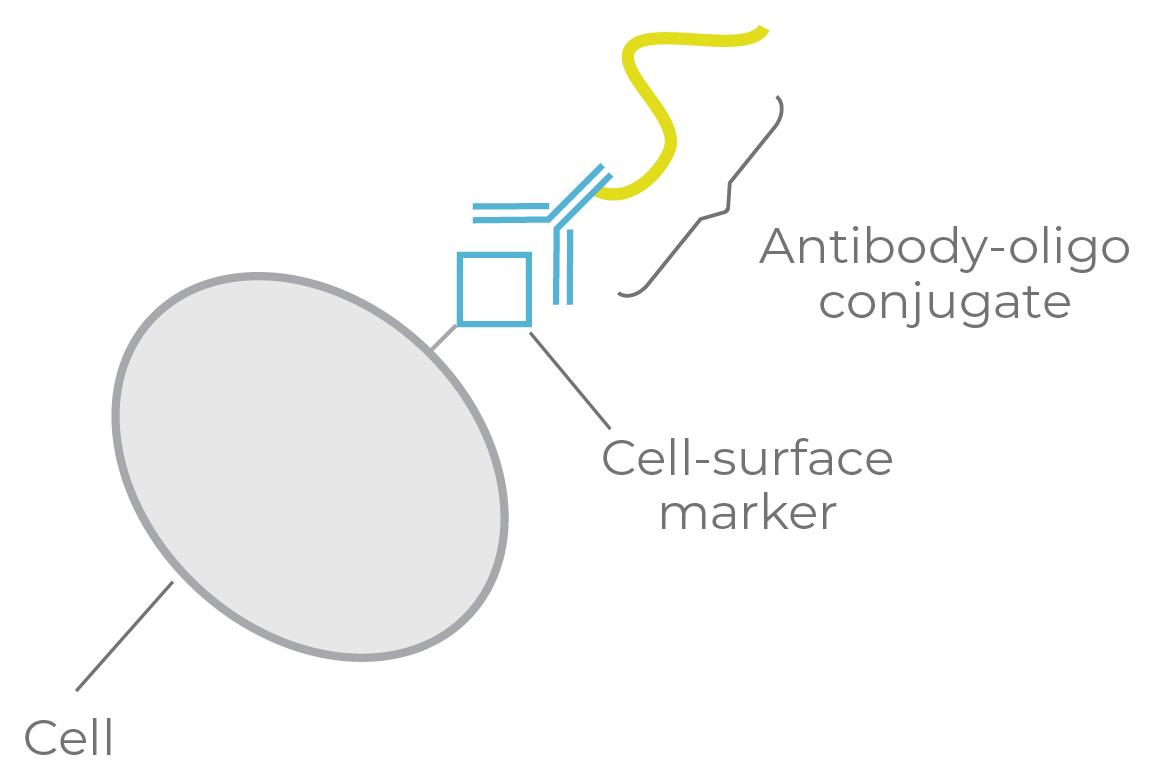
Figure 9a. Single-cell DNA + protein targeting. Antibody-oligo conjugates are used to tag each cell-surface marker with an oligonucleotide.
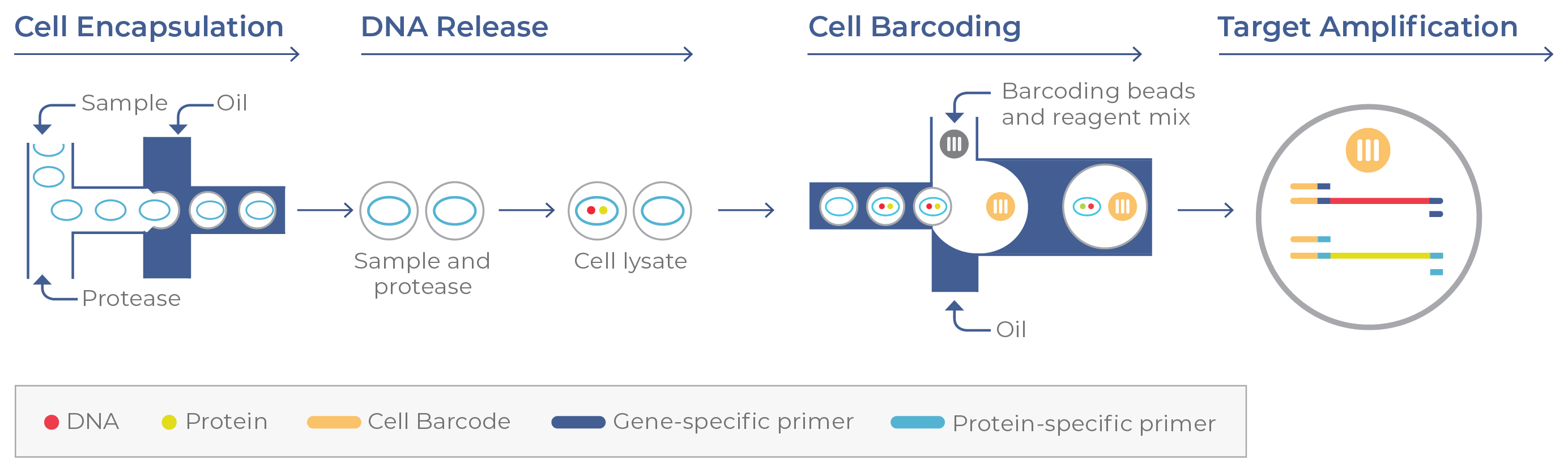
Figure 9b. Single-cell DNA + protein targeting. After cells are stained with a protein panel, the sample is deposited in a Tapestri cartridge, where individual cells are encapsulated in oil droplets with a protease (releases the DNA). Targeted DNA and protein-specific oligos are tagged with a cell-specific barcode during target amplification.
Single-cell multi-omics analyses on Tapestri yield rich datasets that enable robust cellular classifications based on genotypic and immunophenotypic profiles. The dataset in Figure 10 depicts distinct groupings of four cell lines based on SNV, CNV, and protein information. The plot that utilizes all three parameters portrays the most distinct clusters (far right). This example illustrates how multi-modal analyses can reveal classifications that are not apparent with a single analyte.
As exemplified here, single-cell multi-omics and DNA sequencing is enabling cancer to be scrutinized with an unprecedented level of resolution. Oncologists are quickly capitalizing on the use of these exciting technologies to interrogate various heme malignancies and solid tumors.4-8 Next, we explore the use of single-cell DNA and multi-omics in cancer research, with a particular focus on translational applications for the clinic.
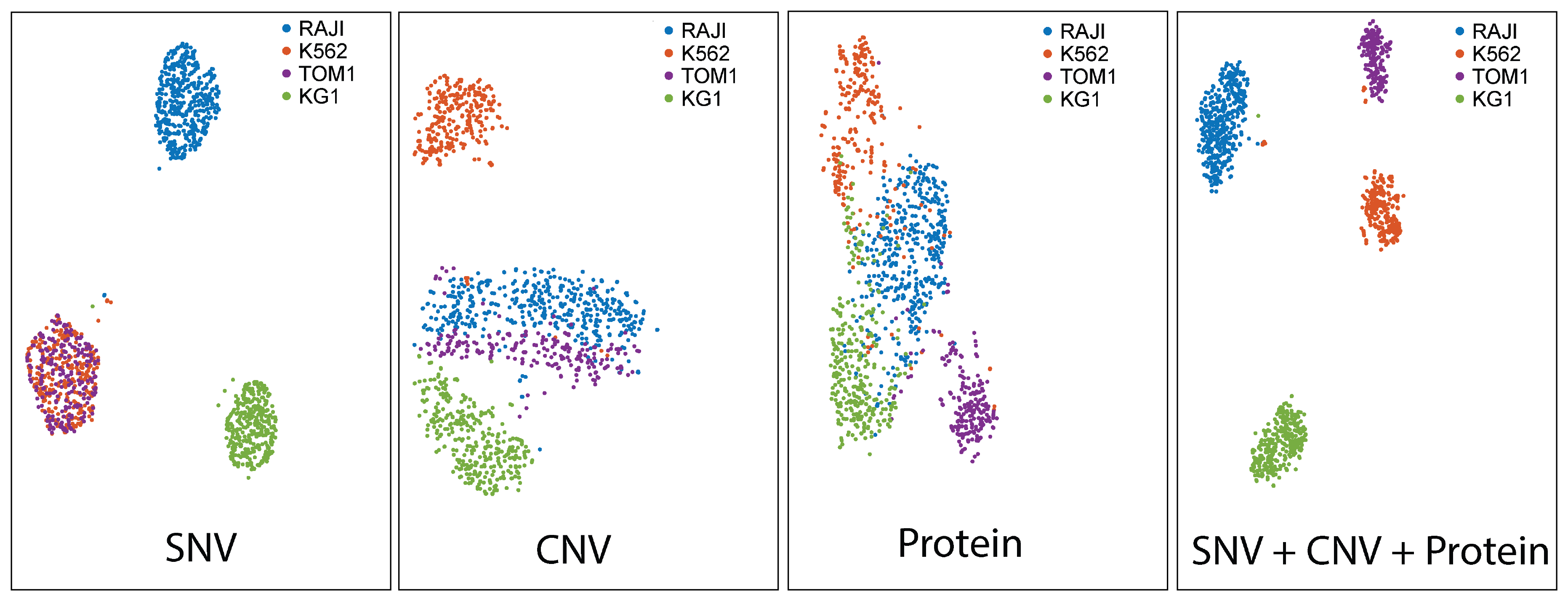
Figure 10. Single-cell multi-omics resolves distinct cell populations. t-SNE projections of single-cell DNA + protein data showing clustering of RAJI, K562, TOM1, and KG1 cell lines. Note that all combining genotype and protein information yielded the most distinct clusters (blue box).
Bulk next-generation sequencing (bulk NGS): a technique where pooled DNA is sequenced and averaged across cells.
Variant allele frequency (VAF): the percentage of mutant alleles in the sample (the proportion of reads that match a particular gene variant divided by the coverage at the locus).
Single-cell DNA sequencing: Cell-specific DNA targeting and NGS.
Tapestri Platform: a single-cell DNA and multi-omics platform by Mission Bio.
Single-cell multi-omics: the process in which multiple analytes are measured from single cells, such as DNA, mRNA, and protein (or any combination of these).
Immunophenotype: a cell’s profile of cell-surface markers.
Proteogenomic profiling: the simultaneous measurement of DNA and protein (in this case cell-surface markers).
Antibody-oligo conjugate (AOC): an antibody linked to a short oligonucleotide.
